Standardization of 16S rRNA gene sequencing using nanopore long read sequencing technology for clinical diagnosis of culture negative infections
- PMID: 40115075
- PMCID: PMC11922894
- DOI: 10.3389/fcimb.2025.1517208
Standardization of 16S rRNA gene sequencing using nanopore long read sequencing technology for clinical diagnosis of culture negative infections
Abstract
The integration of long-read sequencing technology, such as nanopore sequencing technology [Oxford Nanopore Technologies (ONT)], into routine diagnostic laboratories has the potential to transform bacterial infection diagnostics and improve patient management. Analysis of amplicons from long-read sequencing of the 16S rRNA gene generates a comprehensive view of the microbial community within clinical samples, significantly enhancing sensitivity and capacity to analyse mixed bacterial populations compared to short read sequencing approaches. This study evaluates various ONT sequencing approaches and library preparation kits to establish a reliable testing and quality framework for clinical implementation. This study highlights the critical importance of using well-characterized reference materials in validating and revalidating long-read sequencing methods, leveraging a combination of standardized reference materials and clinical samples to navigate the evolving landscape of microbial diagnostics. It presents a robust validation framework for laboratory accreditation and outlines a methodology for comparing the performance of newer ONT chemistries with earlier versions. Additionally, the study details the methods and quality control measures necessary for achieving more accurate and efficient diagnoses of bacterial infections, ultimately reducing time to treatment and enhancing patient outcomes.
Keywords: 16S rRNA; Oxford nanopore; culture negative; long read sequencing; standardization.
Copyright © 2025 Butler, Turner, Mohammed, Akhtar, Evans, Lambourne, Harris, O'Sullivan and Sergaki.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
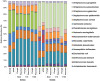

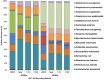

References
-
- Anwar S., Mate R., Preni Sinnakandu L. H., Tierney S., Carmelina, Vinci P. M., Malik K., et al. . (2023). A who collaborative study to evaluate the candidate who international reference reagent for dna extraction of the gut microbiome (World Health Organization - Expert Committee On Biological Standardization; ). Available at: https://www.who.int/publications/m/item/who-bs-2022.2416.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

