Construction and validation of a nomogram model for predicting peritoneal metastasis in gastric cancer based on ferroptosis-relate genes and clinicopathological features
- PMID: 40115916
- PMCID: PMC11921409
- DOI: 10.21037/jgo-24-670
Construction and validation of a nomogram model for predicting peritoneal metastasis in gastric cancer based on ferroptosis-relate genes and clinicopathological features
Abstract
Background: Gastric cancer peritoneal metastasis (GCPM) is a lethal condition. Current diagnostic methods for GCPM, such as imaging and serum tumor markers, lack specificity and sensitivity. Research suggests that utilizing gene signatures to predict GCPM shows significant predictive ability. Nonetheless, the predictability of GCPM using ferroptosis-related genes (FRGs) remains unknown. We aim to construct a nomogram based on FRGs for early diagnosis of GCPM.
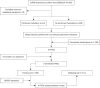
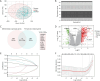
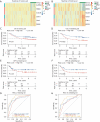
Methods: RNA sequencing and clinical data of patients with gastric cancer (GC) were downloaded from Gene Expression Omnibus (GEO) databases. GCPM was diagnosed through imaging, biopsy and cytology. A GCPM prediction model was developed based on six distinctively expressed FRGs, and the efficiency of the model was assessed through receiver operating characteristic (ROC) curves in both experimental and validation cohorts. Subsequently, 115 clinical samples were examined by immunohistochemistry (IHC) to validate the prediction model's accuracy.
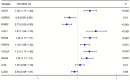
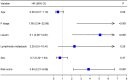
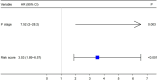
Results: Our analysis included 282 patients, among whom 54 had GCPM while 228 did not. Patients were randomly distributed into experimental and validation groups at a 3:2 ratio. Least absolute shrinkage and selection operator (LASSO) regression identified the coefficients of six FRGs, with a risk score calculated for every patient. Univariate and multivariate logistic analyses revealed that both risk score and pathological stage were significantly associated with GCPM. The area under the curve (AUC) values for the training and validating sets implied good predictability for GCPM (0.827 and 0.767, respectively). Combining the risk score with the tumor node metastasis (TNM) stage substantially improved predictability (AUCs were 0.916 and 0.848 respectively). Lastly, a nomogram incorporating the risk score and TNM stage was constructed, which shows good clinical utility through decision curve analysis (DCA). The IHC results from 115 clinical samples were consistent with these findings.
Conclusions: A nomogram model based on FRGs and clinicopathological features was constructed, demonstrating impressive predictive value for GCPM. This enables timely and personalized therapeutic interventions, thereby benefiting gastric cancer patients.
Keywords: Gastric cancer (GC); RRM2; ferroptosis; peritoneal metastasis; predictive signature.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-670/coif). The authors have no conflicts of interest to declare.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous
