Identification and expression responses of TCP gene family in Opisthopappus taihangensis under abiotic stress
- PMID: 40115945
- PMCID: PMC11922953
- DOI: 10.3389/fpls.2025.1499244
Identification and expression responses of TCP gene family in Opisthopappus taihangensis under abiotic stress
Abstract
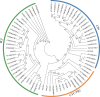
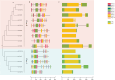
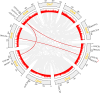
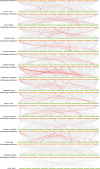
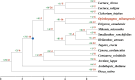
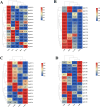
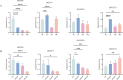
The TCP gene family plays pivotal roles in the development and abiotic stress responses of plants; however, no data has been provided for this gene family in Opisthopappus taihangensis. Based on O. taihangensis genome, 14 TCP genes were identified and divided into two classes (I and II). After tandem and segmental duplication/whole-genome duplication (WGD), more loss and less gain events of OtTCPs occurred, which might be related with the underwent purifying selection during the evolution. The conserved motifs and structures of OtTCP genes contained light response, growth and development, hormone response, and stress-related cis-acting elements. Different OtTCP genes, even duplicated gene pairs, could be expressed in different tissues, which implied that OtTCP genes had diverse function. Among OtTCPs, OtTCP4, 9 and 11 of CYC clade (Class II) presented a relative wide expression pattern with no or one intron. The three TCP genes could be regarded as important candidate factors for O. taihangensis in growth, development and stress response. These results provided some clues and references for the further in-depth exploration of O. taihangensis resistance mechanisms, as well as those of other unique eco-environment plants.
Keywords: Opisthopappus taihangensis; TCP gene family; abiotic stress; evolution; gene expression analysis.
Copyright © 2025 Gao, Zhou, Han, Shen, Zhang, Wu, Dan, Wang, Ye, Liu, Chai and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Potential Response Patterns of Endogenous Hormones in Cliff Species Opisthopappus taihangensis and Opisthopappus longilobus under Salt Stress.Plants (Basel). 2024 Feb 19;13(4):557. doi: 10.3390/plants13040557. Plants (Basel). 2024. PMID: 38498538 Free PMC article.
-
Genome-wide identification, characterization and expression of C2H2 zinc finger gene family in Opisthopappus species under salt stress.BMC Genomics. 2024 Apr 19;25(1):385. doi: 10.1186/s12864-024-10273-7. BMC Genomics. 2024. PMID: 38641598 Free PMC article.
-
Genome-wide identification of oat TCP gene family and expression patterns under abiotic stress.Front Genet. 2025 Feb 4;16:1533562. doi: 10.3389/fgene.2025.1533562. eCollection 2025. Front Genet. 2025. PMID: 39967685 Free PMC article.
-
Physiological and transcriptome analyses of Opisthopappus taihangensis in response to drought stress.Cell Biosci. 2019 Jul 4;9:56. doi: 10.1186/s13578-019-0318-7. eCollection 2019. Cell Biosci. 2019. PMID: 31312427 Free PMC article.
-
Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology.Front Genet. 2022 Nov 9;13:1060546. doi: 10.3389/fgene.2022.1060546. eCollection 2022. Front Genet. 2022. PMID: 36437962 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

