Microbes display broad diversity in cobamide preferences
- PMID: 40116488
- PMCID: PMC12013260
- DOI: 10.1128/msystems.01407-24
Microbes display broad diversity in cobamide preferences
Abstract
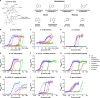
Cobamides, the vitamin B12 (cobalamin) family of cofactors, are used by most organisms but produced by only a fraction of prokaryotes, and are thus considered key shared nutrients among microbes. Cobamides are structurally diverse, with multiple different cobamides found in most microbial communities. The ability to use different cobamides has been tested for several bacteria and microalgae, and nearly all show preferences for certain cobamides. This approach is limited by the commercial unavailability of cobamides other than cobalamin. Here, we have extracted and purified seven commercially unavailable cobamides to characterize bacterial cobamide preferences based on growth in specific cobamide-dependent conditions. The tested bacteria include engineered strains of Escherichia coli, Sinorhizobium meliloti, and Bacillus subtilis expressing native or heterologous cobamide-dependent enzymes, cultured under conditions that functionally isolate specific cobamide-dependent processes such as methionine synthesis. Comparison of these results to those of previous studies of diverse bacteria and microalgae revealed that a broad diversity of cobamide preferences exists not only across different organisms but also between different cobamide-dependent metabolic pathways within the same organism. The microbes differed in the cobamides that support growth most efficiently, cobamides that do not support growth, and the minimum cobamide concentrations required for growth. The latter differ by up to four orders of magnitude across organisms from different environments and by up to 20-fold between cobamide-dependent enzymes within the same organism. Given that cobamides are shared, required for use of specific growth substrates, and essential for central metabolism in certain organisms, cobamide preferences likely impact community structure and function.IMPORTANCENearly all bacteria are found in microbial communities with tens to thousands of other species. Molecular interactions such as metabolic cooperation and competition are key factors underlying community assembly and structure. Cobamides, the vitamin B12 family of enzyme cofactors, are one such class of nutrients, produced by only a minority of prokaryotes but required by most microbes. A unique aspect of cobamides is their broad diversity, with nearly 20 structural forms identified in nature. Importantly, this structural diversity impacts growth as most bacteria that have been tested show preferences for specific cobamide forms. We measured cobamide-dependent growth in several model bacteria and compared the results to those of previous analyses of cobamide preference. We found that cobamide preferences vary widely across bacteria, showing the importance of characterizing these aspects of cobamide biology to understand the impact of cobamides on microbial communities.
Keywords: cobalamin; cobamide; cobamide preference; corrinoid; vitamin B12.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Update of
-
Microbes display broad diversity in cobamide preferences.bioRxiv [Preprint]. 2025 Feb 20:2024.11.04.621602. doi: 10.1101/2024.11.04.621602. bioRxiv. 2025. Update in: mSystems. 2025 Apr 22;10(4):e0140724. doi: 10.1128/msystems.01407-24. PMID: 39574663 Free PMC article. Updated. Preprint.
Similar articles
-
Microbes display broad diversity in cobamide preferences.bioRxiv [Preprint]. 2025 Feb 20:2024.11.04.621602. doi: 10.1101/2024.11.04.621602. bioRxiv. 2025. Update in: mSystems. 2025 Apr 22;10(4):e0140724. doi: 10.1128/msystems.01407-24. PMID: 39574663 Free PMC article. Updated. Preprint.
-
Laboratory evolution of E. coli with a natural vitamin B12 analog reveals roles for cobamide uptake and adenosylation in methionine synthase-dependent growth.J Bacteriol. 2025 Feb 20;207(2):e0028424. doi: 10.1128/jb.00284-24. Epub 2025 Jan 28. J Bacteriol. 2025. PMID: 39873498 Free PMC article.
-
Cofactor Selectivity in Methylmalonyl Coenzyme A Mutase, a Model Cobamide-Dependent Enzyme.mBio. 2019 Sep 24;10(5):e01303-19. doi: 10.1128/mBio.01303-19. mBio. 2019. PMID: 31551329 Free PMC article.
-
Comparative Analysis of Corrinoid Profiles across Host-Associated and Environmental Samples.Biochemistry. 2022 Dec 20;61(24):2791-2796. doi: 10.1021/acs.biochem.2c00367. Epub 2022 Aug 29. Biochemistry. 2022. PMID: 36037062 Review.
-
Sharing vitamins: Cobamides unveil microbial interactions.Science. 2020 Jul 3;369(6499):eaba0165. doi: 10.1126/science.aba0165. Science. 2020. PMID: 32631870 Free PMC article. Review.
References
-
- Renz P. 1999. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids, p 557–566. In Banerjee R (ed), Chemistry and biochemistry of B12. John Wiley & Sons, Inc, New York.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

