SARS-CoV-2 cellular coinfection is limited by superinfection exclusion
- PMID: 40116503
- PMCID: PMC11998510
- DOI: 10.1128/jvi.02077-24
SARS-CoV-2 cellular coinfection is limited by superinfection exclusion
Abstract
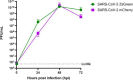
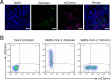
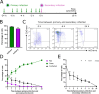
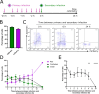
The coinfection of individual cells is a requirement for exchange between two or more virus genomes, which is a major mechanism driving virus evolution. Coinfection is restricted by a mechanism known as superinfection exclusion (SIE), which prohibits the infection of a previously infected cell by a related virus after a period of time. SIE regulates coinfection for many different viruses, but its relevance to the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was unknown. In this study, we investigated this using a pair of SARS-CoV-2 variant viruses encoding distinct fluorescent reporter proteins. We show for the first time that SARS-CoV-2 coinfection of individual cells is limited temporally by SIE. We defined the kinetics of the onset of SIE for SARS-CoV-2 in this system, showing that the potential for coinfection starts to diminish within the first hour of primary infection and then falls exponentially as the time between the two infection events is increased. We then asked how these kinetics would affect the potential for coinfection with viruses during a spreading infection. We used plaque assays to model the localized spread of SARS-CoV-2 observed in infected tissue and showed that the kinetics of SIE restrict coinfection-and therefore sites of possible genetic exchange-to a small interface of infected cells between spreading viral infections. This indicates that SIE, by reducing the likelihood of coinfection of cells, likely reduces the opportunities for genetic exchange between different strains of SARS-CoV-2 and therefore is an underappreciated factor in shaping SARS-CoV-2 evolution.
Importance: Since SARS-CoV-2 first emerged in 2019, it has continued to evolve, occasionally generating variants of concern. One of the ways that SARS-CoV-2 can evolve is through recombination, where genetic information is swapped between different genomes. Recombination requires the coinfection of cells; therefore, factors impacting coinfection are likely to influence SARS-CoV-2 evolution. Coinfection is restricted by SIE, a phenomenon whereby a previously infected cell becomes increasingly resistant to subsequent infection. Here we report that SIE is activated following SARS-CoV-2 infection and reduces the likelihood of coinfection exponentially following primary infection. Furthermore, we show that SIE prevents coinfection of cells at the boundary between two expanding areas of infection, the scenario most likely to lead to recombination between different SARS-CoV-2 lineages. Our work suggests that SIE reduces the likelihood of recombination between SARS-CoV-2 genomes and therefore likely shapes SARS-CoV-2 evolution.
Keywords: SARS-CoV-2; coinfection; coronavirus; superinfection exclusion (SIE); virus-virus interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Superinfection exclusion creates spatially distinct influenza virus populations.PLoS Biol. 2023 Feb 9;21(2):e3001941. doi: 10.1371/journal.pbio.3001941. eCollection 2023 Feb. PLoS Biol. 2023. PMID: 36757937 Free PMC article.
-
A viral protein mediates superinfection exclusion at the whole-organism level but is not required for exclusion at the cellular level.J Virol. 2014 Oct;88(19):11327-38. doi: 10.1128/JVI.01612-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031351 Free PMC article.
-
Unusual SARS-CoV-2 intrahost diversity reveals lineage superinfection.Microb Genom. 2022 Mar;8(3):000751. doi: 10.1099/mgen.0.000751. Microb Genom. 2022. PMID: 35297757 Free PMC article.
-
Chikungunya virus superinfection exclusion is mediated by a block in viral replication and does not rely on non-structural protein 2.PLoS One. 2020 Nov 12;15(11):e0241592. doi: 10.1371/journal.pone.0241592. eCollection 2020. PLoS One. 2020. PMID: 33180795 Free PMC article.
-
The Prevalence and Impact of Coinfection and Superinfection on the Severity and Outcome of COVID-19 Infection: An Updated Literature Review.Pathogens. 2022 Apr 7;11(4):445. doi: 10.3390/pathogens11040445. Pathogens. 2022. PMID: 35456120 Free PMC article. Review.
Cited by
-
Induction of tunnelling nanotube-like structures by influenza A viruses requires the onset of apoptosis.PLoS Pathog. 2025 Jun 5;21(6):e1013191. doi: 10.1371/journal.ppat.1013191. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40472024 Free PMC article.
-
The fitness consequences of coinfection and reassortment for segmented viruses depend upon viral genetic structure.bioRxiv [Preprint]. 2025 Jul 26:2025.07.22.666171. doi: 10.1101/2025.07.22.666171. bioRxiv. 2025. PMID: 40777271 Free PMC article. Preprint.
References
-
- WHO . 2022. Interactive Timeline: WHO’s COVID-19 Response. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interact...
-
- WHO . 2025. WHO COVID-19 Dashboard. https://covid19.who.int/.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

