A metabolic shift to the serine pathway induced by lipids fosters epigenetic reprogramming in nontransformed breast cells
- PMID: 40117373
- PMCID: PMC11927636
- DOI: 10.1126/sciadv.ads9182
A metabolic shift to the serine pathway induced by lipids fosters epigenetic reprogramming in nontransformed breast cells
Abstract
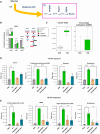
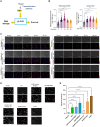
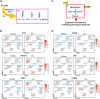
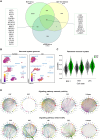
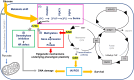
Lipid metabolism and the serine, one-carbon, glycine (SOG) and methionine pathways are independently and significantly correlated with estrogen receptor-negative breast cancer (ERneg BC). Here, we propose a link between lipid metabolism and ERneg BC through phosphoglycerate dehydrogenase (PHGDH), the rate-limiting enzyme in the de novo serine pathway. We demonstrate that the metabolism of the paradigmatic medium-chain fatty acid octanoic acid leads to a metabolic shift toward the SOG and methionine pathways. PHGDH plays a role in both the forward direction, contributing to the production of S-adenosylmethionine, and the reverse direction, generating the oncometabolite 2-hydroxyglutarate, leading to epigenomic reprogramming and phenotypic plasticity. The methionine cycle is closely linked to the transsulfuration pathway. Consequently, we observe that the shift increases the antioxidant glutathione, which mitigates reactive oxygen species (ROS), enabling survival of a subset of cells that have undergone DNA damage. These metabolic changes contribute to several hallmarks of cancer.
Figures



References
-
- Global Burden of Disease 2019 Cancer Collaboration , Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: A systematic analysis for the global burden of disease study 2019. JAMA Oncol. 8, 420–444 (2022). - PMC - PubMed
-
- Sporn M. B., Approaches to prevention of epithelial cancer during the preneoplastic period. Cancer Res. 36, 2699–2702 (1976). - PubMed
-
- Yang X. R., Chang-Claude J., Goode E. L., Couch F. J., Nevanlinna H., Milne R. L., Gaudet M., Schmidt M. K., Broeks A., Cox A., Fasching P. A., Hein R., Spurdle A. B., Blows F., Driver K., Flesch-Janys D., Heinz J., Sinn P., Vrieling A., Heikkinen T., Aittomäki K., Heikkilä P., Blomqvist C., Lissowska J., Peplonska B., Chanock S., Figueroa J., Brinton L., Hall P., Czene K., Humphreys K., Darabi H., Liu J., 't Veer L. J., Leeuwen F. E., Andrulis I. L., Glendon G., Knight J. A., Mulligan A. M., O'Malley F. P., Weerasooriya N., John E. M., Beckmann M. W., Hartmann A., Weihbrecht S. B., Wachter D. L., Jud S. M., Loehberg C. R., Baglietto L., English D. R., Giles G. G., McLean C. A., Severi G., Lambrechts D., Vandorpe T., Weltens C., Paridaens R., Smeets A., Neven P., Wildiers H., Wang X., Olson J. E., Cafourek V., Fredericksen Z., Kosel M., Vachon C., Cramp H. E., Connley D., Cross S. S., Balasubramanian S. P., Reed M. W., Dörk T., Bremer M., Meyer A., Karstens J. H., Ay A., Park-Simon T. W., Hillemanns P., Arias Pérez J. I., Menéndez Rodríguez P., Zamora P., Benítez J., Ko Y. D., Fischer H. P., Hamann U., Pesch B., Brüning T., Justenhoven C., Brauch H., Eccles D. M., Tapper W. J., Gerty S. M., Sawyer E. J., Tomlinson I. P., Jones A., Kerin M., Miller N., McInerney N., Anton-Culver H., Ziogas A., Shen C. Y., Hsiung C. N., Wu P. E., Yang S. L., Yu J. C., Chen S. T., Hsu G. C., Haiman C. A., Henderson B. E., Marchand L., Kolonel L. N., Lindblom A., Margolin S., Jakubowska A., Lubiński J., Huzarski T., Byrski T., Górski B., Gronwald J., Hooning M. J., Hollestelle A., Ouweland A. M., Jager A., Kriege M., Tilanus-Linthorst M. M., Collée M., Wang-Gohrke S., Pylkäs K., Jukkola-Vuorinen A., Mononen K., Grip M., Hirvikoski P., Winqvist R., Mannermaa A., Kosma V. M., Kauppinen J., Kataja V., Auvinen P., Soini Y., Sironen R., Bojesen S. E., Ørsted D. D., Kaur-Knudsen D., Flyger H., Nordestgaard B. G., Holland H., Chenevix-Trench G., Manoukian S., Barile M., Radice P., Hankinson S. E., Hunter D. J., Tamimi R., Sangrajrang S., Brennan P., McKay J., Odefrey F., Gaborieau V., Devilee P., Huijts P. E., Tollenaar R. A., Seynaeve C., Dite G. S., Apicella C., Hopper J. L., Hammet F., Tsimiklis H., Smith L. D., Southey M. C., Humphreys M. K., Easton D., Pharoah P., Sherman M. E., Garcia-Closas M., Associations of breast cancer risk factors with tumor subtypes: A pooled analysis from the Breast Cancer Association Consortium studies. J. Natl. Cancer Inst. 103, 250–263 (2011). - PMC - PubMed
-
- Milne R. L., Kuchenbaecker K. B., Michailidou K., Beesley J., Kar S., Lindström S., Hui S., Lemaçon A., Soucy P., Dennis J., Jiang X., Rostamianfar A., Finucane H., Bolla M. K., McGuffog L., Wang Q., Aalfs C. M., Adams M., Adlard J., Agata S., Ahmed S., Ahsan H., Aittomäki K., Al-Ejeh F., Allen J., Ambrosone C. B., Amos C. I., Andrulis I. L., Anton-Culver H., Antonenkova N. N., Arndt V., Arnold N., Aronson K. J., Auber B., Auer P. L., Ausems M., Azzollini J., Bacot F., Balmaña J., Barile M., Barjhoux L., Barkardottir R. B., Barrdahl M., Barnes D., Barrowdale D., Baynes C., Beckmann M. W., Benitez J., Bermisheva M., Bernstein L., Bignon Y. J., Blazer K. R., Blok M. J., Blomqvist C., Blot W., Bobolis K., Boeckx B., Bogdanova N. V., Bojesen A., Bojesen S. E., Bonanni B., Børresen-Dale A. L., Bozsik A., Bradbury A. R., Brand J. S., Brauch H., Brenner H., Paillerets B. B.-d., Brewer C., Brinton L., Broberg P., Brooks-Wilson A., Brunet J., Brüning T., Burwinkel B., Buys S. S., Byun J., Cai Q., Caldés T., Caligo M. A., Campbell I., Canzian F., Caron O., Carracedo A., Carter B. D., Castelao J. E., Castera L., Caux-Moncoutier V., Chan S. B., Chang-Claude J., Chanock S. J., Chen X., Cheng T. D., Chiquette J., Christiansen H., Claes K. B. M., Clarke C. L., Conner T., Conroy D. M., Cook J., Cordina-Duverger E., Cornelissen S., Coupier I., Cox A., Cox D. G., Cross S. S., Cuk K., Cunningham J. M., Czene K., Daly M. B., Damiola F., Darabi H., Davidson R., De Leeneer K., Devilee P., Dicks E., Diez O., Ding Y. C., Ditsch N., Doheny K. F., Domchek S. M., Dorfling C. M., Dörk T., Dos-Santos-Silva I., Dubois S., Dugué P. A., Dumont M., Dunning A. M., Durcan L., Dwek M., Dworniczak B., Eccles D., Eeles R., Ehrencrona H., Eilber U., Ejlertsen B., Ekici A. B., Eliassen A. H., Engel C., Eriksson M., Fachal L., Faivre L., Fasching P. A., Faust U., Figueroa J., Flesch-Janys D., Fletcher O., Flyger H., Foulkes W. D., Friedman E., Fritschi L., Frost D., Gabrielson M., Gaddam P., Gammon M. D., Ganz P. A., Gapstur S. M., Garber J., Garcia-Barberan V., García-Sáenz J. A., Gaudet M. M., Gauthier-Villars M., Gehrig A., Georgoulias V., Gerdes A. M., Giles G. G., Glendon G., Godwin A. K., Goldberg M. S., Goldgar D. E., González-Neira A., Goodfellow P., Greene M. H., Alnæs G. I. G., Grip M., Gronwald J., Grundy A., Gschwantler-Kaulich D., Guénel P., Guo Q., Haeberle L., Hahnen E., Haiman C. A., Håkansson N., Hallberg E., Hamann U., Hamel N., Hankinson S., Hansen T. V. O., Harrington P., Hart S. N., Hartikainen J. M., Healey C. S., Hein A., Helbig S., Henderson A., Heyworth J., Hicks B., Hillemanns P., Hodgson S., Hogervorst F. B., Hollestelle A., Hooning M. J., Hoover B., Hopper J. L., Hu C., Huang G., Hulick P. J., Humphreys K., Hunter D. J., Imyanitov E. N., Isaacs C., Iwasaki M., Izatt L., Jakubowska A., James P., Janavicius R., Janni W., Jensen U. B., John E. M., Johnson N., Jones K., Jones M., Jukkola-Vuorinen A., Kaaks R., Kabisch M., Kaczmarek K., Kang D., Kast K., Keeman R., Kerin M. J., Kets C. M., Keupers M., Khan S., Khusnutdinova E., Kiiski J. I., Kim S. W., Knight J. A., Konstantopoulou I., Kosma V. M., Kristensen V. N., Kruse T. A., Kwong A., Lænkholm A. V., Laitman Y., Lalloo F., Lambrechts D., Landsman K., Lasset C., Lazaro C., Le Marchand L., Lecarpentier J., Lee A., Lee E., Lee J. W., Lee M. H., Lejbkowicz F., Lesueur F., Li J., Lilyquist J., Lincoln A., Lindblom A., Lissowska J., Lo W. Y., Loibl S., Long J., Loud J. T., Lubinski J., Luccarini C., Lush M., MacInnis R. J., Maishman T., Makalic E., Kostovska I. M., Malone K. E., Manoukian S., Manson J. E., Margolin S., Martens J. W. M., Martinez M. E., Matsuo K., Mavroudis D., Mazoyer S., McLean C., Meijers-Heijboer H., Menéndez P., Meyer J., Miao H., Miller A., Miller N., Mitchell G., Montagna M., Muir K., Mulligan A. M., Mulot C., Nadesan S., Nathanson K. L., Neuhausen S. L., Nevanlinna H., Nevelsteen I., Niederacher D., Nielsen S. F., Nordestgaard B. G., Norman A., Nussbaum R. L., Olah E., Olopade O. I., Olson J. E., Olswold C., Ong K. R., Oosterwijk J. C., Orr N., Osorio A., Pankratz V. S., Papi L., Park-Simon T. W., Paulsson-Karlsson Y., Lloyd R., Pedersen I. S., Peissel B., Peixoto A., Perez J. I. A., Peterlongo P., Peto J., Pfeiler G., Phelan C. M., Pinchev M., Plaseska-Karanfilska D., Poppe B., Porteous M. E., Prentice R., Presneau N., Prokofieva D., Pugh E., Pujana M. A., Pylkäs K., Rack B., Radice P., Rahman N., Rantala J., Rappaport-Fuerhauser C., Rennert G., Rennert H. S., Rhenius V., Rhiem K., Richardson A., Rodriguez G. C., Romero A., Romm J., Rookus M. A., Rudolph A., Ruediger T., Saloustros E., Sanders J., Sandler D. P., Sangrajrang S., Sawyer E. J., Schmidt D. F., Schoemaker M. J., Schumacher F., Schürmann P., Schwentner L., Scott C., Scott R. J., Seal S., Senter L., Seynaeve C., Shah M., Sharma P., Shen C. Y., Sheng X., Shimelis H., Shrubsole M. J., Shu X. O., Side L. E., Singer C. F., Sohn C., Southey M. C., Spinelli J. J., Spurdle A. B., Stegmaier C., Stoppa-Lyonnet D., Sukiennicki G., Surowy H., Sutter C., Swerdlow A., Szabo C. I., Tamimi R. M., Tan Y. Y., Taylor J. A., Tejada M. I., Tengström M., Teo S. H., Terry M. B., Tessier D. C., Teulé A., Thöne K., Thull D. L., Tibiletti M. G., Tihomirova L., Tischkowitz M., Toland A. E., Tollenaar R., Tomlinson I., Tong L., Torres D., Tranchant M., Truong T., Tucker K., Tung N., Tyrer J., Ulmer H. U., Vachon C., van Asperen C. J., Van Den Berg D., van den Ouweland A. M. W., van Rensburg E. J., Varesco L., Varon-Mateeva R., Vega A., Viel A., Vijai J., Vincent D., Vollenweider J., Walker L., Wang Z., Wang-Gohrke S., Wappenschmidt B., Weinberg C. R., Weitzel J. N., Wendt C., Wesseling J., Whittemore A. S., Wijnen J. T., Willett W., Winqvist R., Wolk A., Wu A. H., Xia L., Yang X. R., Yannoukakos D., Zaffaroni D., Zheng W., Zhu B., Ziogas A., Ziv E., Zorn K. K., Gago-Dominguez M., Mannermaa A., Olsson H., Teixeira M. R., Stone J., Offit K., Ottini L., Park S. K., Thomassen M., Hall P., Meindl A., Schmutzler R. K., Droit A., Bader G. D., Pharoah P. D. P., Couch F. J., Easton D. F., Kraft P., Chenevix-Trench G., García-Closas M., Schmidt M. K., Antoniou A. C., Simard J., Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 49, 1767–1778 (2017). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

