Chromatin landscape in paired human visceral and subcutaneous adipose tissue and its impact on clinical variables in obesity
- PMID: 40118008
- PMCID: PMC11976249
- DOI: 10.1016/j.ebiom.2025.105653
Chromatin landscape in paired human visceral and subcutaneous adipose tissue and its impact on clinical variables in obesity
Abstract
Background: Obesity is a global health challenge and adipose tissue exhibits distinct depot-specific characteristics impacting differentially on the risk of metabolic comorbidities.
Methods: Here, we integrate chromatin accessibility (ATAC-seq) and gene expression (RNA-seq) data from intra-individually paired human subcutaneous (SAT) and omental visceral adipose tissue (OVAT) samples to unveil depot-specific regulatory mechanisms.
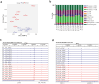
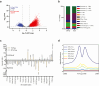
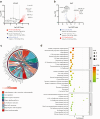
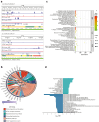
Findings: We identified twice as many depot-specific differentially accessible regions (DARs) in OVAT compared to SAT. SAT-specific regions showed enrichment for adipose tissue enhancers involving genes controlling extracellular matrix organization and metabolic processes. In contrast, OVAT-specific regions showed enrichment in promoters linked to genes associated with cardiomyopathies. Moreover, OVAT-specific regions were enriched for bivalent transcription start site and repressive chromatin states, suggesting a "lingering" regulatory state. Motif analysis identified CTCF and BACH1 as most significantly enriched motifs in SAT and OVAT-specific DARs, respectively. Distinct gene sets correlated with important clinical variables of obesity, fat distribution measures, as well as insulin, glucose, and lipid metabolism.
Interpretation: We provide an integrated analysis of chromatin accessibility and transcriptional profiles in paired human SAT and OVAT samples, offering new insights into the regulatory landscape of adipose tissue and highlighting depot-specific mechanisms in obesity pathogenesis.
Funding: SS received EU-Scientia postdoctoral Fellowship and project funding from the European Union's Horizon 2020 Research and Innovation program under the Marie Skłodowska-Curie Grant, (agreement No. 801133). LlCP and TR were supported by Helse Sør-Øst grants to Y.B (ID 2017079, ID 278908). MB received funding from grants from the DFG (German Research Foundation)-Projekt number 209933838-SFB 1052 (project B1) and by Deutsches Zentrum für Diabetesforschung (DZD, Grant: 82DZD00601).
Keywords: Adipose tissue; Chromatin accessibility; Gene expression; Obesity.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests MB received personal honoraria from Amgen, AstraZeneca, Bayer, Boehringer-Ingelheim, Lilly, Novo Nordisk, Novartis, and Sanofi as well as payments from Boehringer-Ingelheim to the institution. The other authors on the manuscript declared no competing interests.
Figures


References
-
- Bluher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15(5):288–298. - PubMed
-
- Bjorntorp P. Metabolic implications of body fat distribution. Diabetes Care. 1991;14(12):1132–1143. - PubMed
-
- Kissebah A.H., Krakower G.R. Regional adiposity and morbidity. Physiol Rev. 1994;74(4):761–811. - PubMed
-
- Snijder M.B., Visser M., Dekker J.M., et al. Low subcutaneous thigh fat is a risk factor for unfavourable glucose and lipid levels, independently of high abdominal fat. Health ABC Study Diabetol. 2005;48(2):301–308. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

