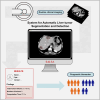
A CT-based deep learning-driven tool for automatic liver tumor detection and delineation in patients with cancer
- PMID: 40118052
- PMCID: PMC12047525
- DOI: 10.1016/j.xcrm.2025.102032
A CT-based deep learning-driven tool for automatic liver tumor detection and delineation in patients with cancer
Abstract
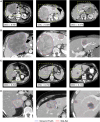
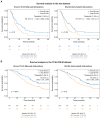
Liver tumors, whether primary or metastatic, significantly impact the outcomes of patients with cancer. Accurate identification and quantification are crucial for effective patient management, including precise diagnosis, prognosis, and therapy evaluation. We present SALSA (system for automatic liver tumor segmentation and detection), a fully automated tool for liver tumor detection and delineation. Developed on 1,598 computed tomography (CT) scans and 4,908 liver tumors, SALSA demonstrates superior accuracy in tumor identification and volume quantification, outperforming state-of-the-art models and inter-reader agreement among expert radiologists. SALSA achieves a patient-wise detection precision of 99.65%, and 81.72% at lesion level, in the external validation cohorts. Additionally, it exhibits good overlap, achieving a dice similarity coefficient (DSC) of 0.760, outperforming both state-of-the-art and the inter-radiologist assessment. SALSA's automatic quantification of tumor volume proves to have prognostic value across various solid tumors (p = 0.028). SALSA's robust capabilities position it as a potential medical device for automatic cancer detection, staging, and response evaluation.
Keywords: deep learning; delineation; detection; imaging; liver tumors.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.P.-L. declares research funding by AstraZeneca and Roche; she participates in the steering committee of a clinical trial sponsored by Roche, not related to this work.
Figures



References
-
- Tsilimigras D.I., Brodt P., Clavien P.-A., Muschel R.J., D’Angelica M.I., Endo I., Parks R.W., Doyle M., de Santibañes E., Pawlik T.M. Liver metastases. Nat. Rev. Dis. Primers. 2021;7:27. - PubMed
-
- Siriwardena A.K., Mason J.M., Mullamitha S., Hancock H.C., Jegatheeswaran S. Management of colorectal cancer presenting with synchronous liver metastases. Nat. Rev. Clin. Oncol. 2014;11:446–459. - PubMed
-
- Yoon S.H., Kim K.W., Goo J.M., Kim D.-W., Hahn S. Observer variability in RECIST-based tumour burden measurements: a meta-analysis. Eur. J. Cancer. 2016;53:5–15. - PubMed
-
- Krasovitsky M., Lee Y.C., Sim H.-W., Chawla T., Moore H., Moses D., Baker L., Mandel C., Kielar A., Hartery A., et al. Interobserver and intraobserver variability of RECIST assessment in ovarian cancer. Int. J. Gynecol. Cancer. 2022;32:656–661. - PubMed

