Binding domain mutations provide insight into CTCF's relationship with chromatin and its contribution to gene regulation
- PMID: 40118069
- PMCID: PMC12008812
- DOI: 10.1016/j.xgen.2025.100813
Binding domain mutations provide insight into CTCF's relationship with chromatin and its contribution to gene regulation
Abstract
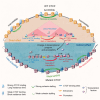
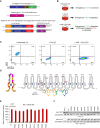
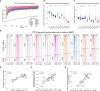
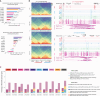
Here we used a series of CTCF mutations to explore CTCF's relationship with chromatin and its contribution to gene regulation. CTCF's impact depends on the genomic context of bound sites and the unique binding properties of WT and mutant CTCF proteins. Specifically, CTCF's signal strength is linked to changes in accessibility, and the ability to block cohesin is linked to its binding stability. Multivariate modeling reveals that both CTCF and accessibility contribute independently to cohesin binding and insulation, but CTCF signal strength has a stronger effect. CTCF and chromatin have a bidirectional relationship such that at CTCF sites, accessibility is reduced in a cohesin-dependent, mutant-specific fashion. In addition, each mutant alters TF binding and accessibility in an indirect manner, changes which impart the most influence on rewiring transcriptional networks and the cell's ability to differentiate. Collectively, the mutant perturbations provide a rich resource for determining CTCF's site-specific effects.
Keywords: CTCF; CTCF mutations; cancer; chromatin accessibility; chromatin organization; cohesin; gene regulation; neurological disorder; residence time.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
Binding domain mutations provide insight into CTCF's relationship with chromatin and its contribution to gene regulation.bioRxiv [Preprint]. 2025 Feb 18:2024.01.11.575070. doi: 10.1101/2024.01.11.575070. bioRxiv. 2025. Update in: Cell Genom. 2025 Apr 9;5(4):100813. doi: 10.1016/j.xgen.2025.100813. PMID: 38370764 Free PMC article. Updated. Preprint.
-
Brain and cancer associated binding domain mutations provide insight into CTCF's relationship with chromatin and its ability to act as a chromatin organizer.Res Sq [Preprint]. 2024 Jul 19:rs.3.rs-4670379. doi: 10.21203/rs.3.rs-4670379/v1. Res Sq. 2024. Update in: Cell Genom. 2025 Apr 9;5(4):100813. doi: 10.1016/j.xgen.2025.100813. PMID: 39070636 Free PMC article. Updated. Preprint.
Similar articles
-
CTCF binding landscape is shaped by the epigenetic state of the N-terminal nucleosome in relation to CTCF motif orientation.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf587. doi: 10.1093/nar/gkaf587. Nucleic Acids Res. 2025. PMID: 40613712 Free PMC article.
-
Binding domain mutations provide insight into CTCF's relationship with chromatin and its contribution to gene regulation.bioRxiv [Preprint]. 2025 Feb 18:2024.01.11.575070. doi: 10.1101/2024.01.11.575070. bioRxiv. 2025. Update in: Cell Genom. 2025 Apr 9;5(4):100813. doi: 10.1016/j.xgen.2025.100813. PMID: 38370764 Free PMC article. Updated. Preprint.
-
High-resolution CTCF footprinting reveals impact of chromatin state on cohesin extrusion.Nat Commun. 2025 May 15;16(1):4506. doi: 10.1038/s41467-025-57775-w. Nat Commun. 2025. PMID: 40374602 Free PMC article.
-
CTCF: the protein, the binding partners, the binding sites and their chromatin loops.Philos Trans R Soc Lond B Biol Sci. 2013 May 6;368(1620):20120369. doi: 10.1098/rstb.2012.0369. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23650640 Free PMC article. Review.
-
On the choreography of genome folding: A grand pas de deux of cohesin and CTCF.Curr Opin Cell Biol. 2021 Jun;70:84-90. doi: 10.1016/j.ceb.2020.12.001. Epub 2021 Feb 2. Curr Opin Cell Biol. 2021. PMID: 33545664 Review.
Cited by
-
Dynamic barriers modulate cohesin positioning and genome folding at fixed occupancy.Genome Res. 2025 Aug 1;35(8):1745-1757. doi: 10.1101/gr.280108.124. Genome Res. 2025. PMID: 40628528 Free PMC article.
-
Wnt signaling activation induces CTCF binding and loop formation at cis-regulatory elements of target genes.Genome Res. 2025 Aug 1;35(8):1701-1716. doi: 10.1101/gr.279684.124. Genome Res. 2025. PMID: 40550689 Free PMC article.
-
CTCF binding landscape is shaped by the epigenetic state of the N-terminal nucleosome in relation to CTCF motif orientation.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf587. doi: 10.1093/nar/gkaf587. Nucleic Acids Res. 2025. PMID: 40613712 Free PMC article.
References
-
- Filippova G.N., Lindblom A., Meincke L.J., Klenova E.M., Neiman P.E., Collins S.J., Doggett N.A., Lobanenkov V.V. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes. Chromosomes. Cancer. 1998;22:26–36. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

