De novo antibody identification in human blood from full-length single B cell transcriptomics and matching haplotype-resolved germline assemblies
- PMID: 40118521
- PMCID: PMC12047243
- DOI: 10.1101/gr.279392.124
De novo antibody identification in human blood from full-length single B cell transcriptomics and matching haplotype-resolved germline assemblies
Abstract
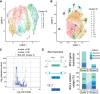
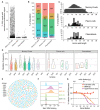
Immunoglobulin (IGH, IGK, IGL) loci in the human genome are highly polymorphic regions that encode the building blocks of the light and heavy chain IG proteins that dimerize to form antibodies. The processes of V(D)J recombination and somatic hypermutation in B cells are responsible for creating an enormous reservoir of highly specific antibodies capable of binding a vast array of possible antigens. However, the antibody repertoire is fundamentally limited by the set of variable (V), diversity (D), and joining (J) alleles present in the germline IG loci. To better understand how the germline IG haplotypes contribute to the expressed antibody repertoire, we combined genome sequencing of the germline IG loci with single-cell transcriptome sequencing of B cells from the same donor. Sequencing and assembly of the germline IG loci captured the IGH locus in a single fully phased contig where the maternal and paternal contributions to the germline V, D, and J repertoire can be fully resolved. The B cells were collected following a measles, mumps, and rubella (MMR) vaccination, resulting in a population of cells that were activated in response to this specific immune challenge. Single-cell, full-length transcriptome sequencing of these B cells results in whole transcriptome characterization of each cell, as well as highly accurate consensus sequences for the somatically rearranged and hypermutated light and heavy chain IG transcripts. A subset of antibodies synthesized based on their consensus heavy and light chain transcript sequences demonstrate binding to measles antigens and neutralization of authentic measles virus.
© 2025 Beaulaurier et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




Update of
-
De novo antibody discovery in human blood from full-length single B cell transcriptomics and matching haplotyped-resolved germline assemblies.bioRxiv [Preprint]. 2024 Mar 29:2024.03.26.586834. doi: 10.1101/2024.03.26.586834. bioRxiv. 2024. Update in: Genome Res. 2025 Apr 14;35(4):929-941. doi: 10.1101/gr.279392.124. PMID: 38585716 Free PMC article. Updated. Preprint.
References
-
- Cho A, Caldara AL, Ran NA, Menne Z, Kauffman RC, Affer M, Llovet A, Norwood C, Scanlan A, Mantus G, et al. 2019. Single-cell analysis suggests that ongoing affinity maturation drives the emergence of pemphigus vulgaris autoimmune disease. Cell Rep 28: 909–922.e6. 10.1016/j.celrep.2019.06.066 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
