Dependence of mitochondrial calcium signalling and dynamics on the disaggregase, CLPB
- PMID: 40118824
- PMCID: PMC11928477
- DOI: 10.1038/s41467-025-57641-9
Dependence of mitochondrial calcium signalling and dynamics on the disaggregase, CLPB
Abstract
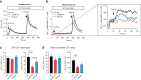
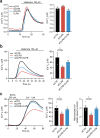
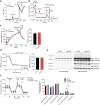
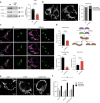
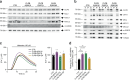
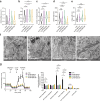
Cells utilize protein disaggregases to avoid abnormal protein aggregation that causes many diseases. Among these, caseinolytic peptidase B protein homolog (CLPB) is localized in the mitochondrial intermembrane space and linked to human disease. Upon CLPB loss, MICU1 and MICU2, regulators of the mitochondrial calcium uniporter complex (mtCU), and OPA1, a main mediator of mitochondrial fusion, become insoluble but the functional outcome remains unclear. In this work we demonstrate that CLPB is required to maintain mitochondrial calcium signalling and fusion dynamics. CLPB loss results in altered mtCU composition, interfering with mitochondrial calcium uptake independently of cytosolic calcium and mitochondrial membrane potential. Additionally, OPA1 decreases, and aggregation occurs, accompanied by mitochondrial fragmentation. Disease-associated mutations in the CLPB gene present in skin fibroblasts from patients also display mitochondrial calcium and structural changes. Thus, mtCU and fusion activity are dependent on CLPB, and their impairments might contribute to the disease caused by CLPB variants.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Louros, N., Schymkowitz, J. & Rousseau, F. Mechanisms and pathology of protein misfolding and aggregation. Nat. Rev. Mol. Cell Biol.24, 912–933 (2023). - PubMed
-
- Capo-Chichi, J. M. et al. Disruption of CLPB is associated with congenital microcephaly, severe encephalopathy and 3-methylglutaconic aciduria. J. Med Genet52, 303–311 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

