Recapitulation of endochondral ossification by hPSC-derived SOX9+ sclerotomal progenitors
- PMID: 40118845
- PMCID: PMC11928506
- DOI: 10.1038/s41467-025-58122-9
Recapitulation of endochondral ossification by hPSC-derived SOX9+ sclerotomal progenitors
Abstract
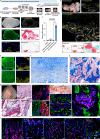
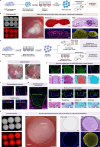
Endochondral ossification generates most of the load-bearing bones, recapitulating it in human cells remains a challenge. Here, we report generation of SOX9+ sclerotomal progenitors (scl-progenitors), a mesenchymal precursor at the pre-condensation stage, from human pluripotent stem cells and development of osteochondral induction methods for these cells. Upon lineage-specific induction, SOX9+ scl-progenitors have not only generated articular cartilage but have also undergone spontaneous condensation, cartilaginous anlagen formation, chondrocyte hypertrophy, vascular invasion, and finally bone formation with stroma, thereby recapitulating key stages during endochondral ossification. Moreover, self-organized growth plate-like structures have also been induced using SOX9+ scl-progenitor-derived fusion constructs with chondro- and osteo-spheroids, exhibiting molecular and cellular similarities to the primary growth plates. Furthermore, we have identified ITGA9 as a specific surface marker for reporter-independent isolation of SOX9+ scl-progenitors and established a culture system to support their expansion. Our work highlights SOX9+ scl-progenitors as a promising tool for modeling human skeletal development and bone/cartilage bioengineering.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Patents for isolation, characterization, and expansion of SOX9+ Scl-progenitors have been filed (application nos. 202211228107.2, 202211220041.2 and 202310160109.0). For all patents, the patent applicant: Sichuan University; name of inventors: Zhonghan Li and Jingfei Xiong; status of application: 202211228107.2 and 202310160109.0 have been granted. All other authors declare no competing interests.
Figures





References
-
- Latourte, A., Kloppenburg, M. & Richette, P. Emerging pharmaceutical therapies for osteoarthritis. Nat. Rev. Rheumatol.16, 673–688 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- 32471507/National Natural Science Foundation of China (National Science Foundation of China)
- 3247120327/National Natural Science Foundation of China (National Science Foundation of China)
- 32071455/National Natural Science Foundation of China (National Science Foundation of China)
- 020SCUNL109/Sichuan University (SCU)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

