Emilia sonchifolia (L.) DC. inhibits the growth of Methicillin-Resistant Staphylococcus epidermidis by modulating its physiology through multiple mechanisms
- PMID: 40119097
- PMCID: PMC11928732
- DOI: 10.1038/s41598-025-93561-w
Emilia sonchifolia (L.) DC. inhibits the growth of Methicillin-Resistant Staphylococcus epidermidis by modulating its physiology through multiple mechanisms
Abstract
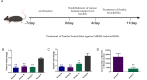
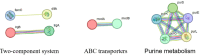
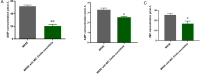
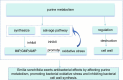
Bloodstream infections (BSIs) are a public health concern, causing substantial morbidity and mortality. Staphylococcus epidermidis (S. epidermidis) is a leading cause BSIs. Antibiotics targeting S. epidermidis have been the mainstay of treatment for BSIs, however their efficacy is diminishing in combating with drug-resistant bacteria. Therefore, alternative treatments for antibiotic-resistant infections are urgently required. Studies have demonstrated that certain traditional Chinese medicine (TCM) exhibit notable antimicrobial activity and can help mitigate bacterial resistance. Among these, The ethanol extract of Emilia sonchifolia (L.) DC (E. sonchifolia) (10 g crude drug/1 g extract ) exhibits a noteworthy anti-methicillin-resistant S. epidermidis (MRSE) effect. This study explores antibacterial activity and underlying mechanisms of E. sonchifolia against MRSE. The antibacterial activity of E. sonchifolia against MRSE was assessed in vitro by measuring the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). The MRSE-induced mouse BSIs model was used to evaluate the antibacterial activity of E. sonchifolia in vivo. Proteomic and transcriptomic analyses were performed to elucidate the underlying antibacterial mechanisms. The MIC and MBC values of E. sonchifolia against MRSE were 5 mg/mL and 20 mg/mL, respectively. In vivo, E. sonchifolia effectively treated MRSE-induced BSIs. Additionally, proteomic and transcriptomic analyses revealed considerable down-regulation of purine metabolism, that were associated with oxidative stress and cell wall synthesis. The enzyme linked immunosorbent assay(ELISA) results showed decreased levels of inosine monophosphate (IMP), Adenosine monophosphate(AMP) and guanine monophosphate (GMP), indicating inhibited purine metabolism. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analysis confirmed bacterial cell wall damage. E. sonchifolia exerts antibacterial effects by inhibiting purine metabolism, promoting bacterial oxidative stress, and impairing cell wall synthesis. These findings provide novel insights into the mechanistic understanding of E. sonchifolia's efficacy against MRSE, offering potential strategies for managing MRSE infections.
Keywords: Emilia sonchifolia; Bloodstream infections; Methicillin-resistant Staphylococcus epidermidis; Purine metabolism.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






References
-
- Lamy, B., Sundqvist, M. & Idelevich, E. A. Bloodstream infections - Standard and progress in pathogen diagnostics. Clin. Microbiol. Infect.26(2), 142–150 (2020). - PubMed
-
- Norris, L. B. et al. Systematic review of antimicrobial lock therapy for prevention of central-line-associated bloodstream infections in adult and pediatric cancer patients. Int. J. Antimicrob. Agents50(3), 308–317 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

