Enhancing human NK cell antitumor function by knocking out SMAD4 to counteract TGFβ and activin A suppression
- PMID: 40119192
- PMCID: PMC11957989
- DOI: 10.1038/s41590-025-02103-z
Enhancing human NK cell antitumor function by knocking out SMAD4 to counteract TGFβ and activin A suppression
Abstract
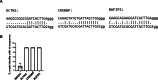
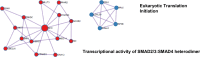
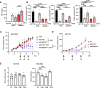
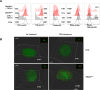
Transforming growth factor beta (TGFβ) and activin A suppress natural killer (NK) cell function and proliferation, limiting the efficacy of adoptive NK cell therapies. Inspired by the partial resistance to TGFβ of NK cells with SMAD4 haploinsufficiency, we used CRISPR-Cas9 for knockout of SMAD4 in human NK cells. Here we show that SMAD4KO NK cells were resistant to TGFβ and activin A inhibition, retaining their cytotoxicity, cytokine secretion and interleukin-2/interleukin-15-driven proliferation. They showed enhanced tumor penetration and tumor growth control, both as monotherapy and in combination with tumor-targeted therapeutic antibodies. Notably, SMAD4KO NK cells outperformed control NK cells treated with a TGFβ inhibitor, underscoring the benefit of maintaining SMAD4-independent TGFβ signaling. SMAD4KO conferred TGFβ resistance across diverse NK cell platforms, including CD19-CAR NK cells, stem cell-derived NK cells and ADAPT-NK cells. These findings position SMAD4 knockout as a versatile and compelling strategy to enhance NK cell antitumor activity, providing a new avenue for improving NK cell-based cancer immunotherapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A. Rea, M.S.-G. and A.M. are inventors of a filed patent (EP23383142, submitted 8 November 2023) pertaining to the results presented in the paper. The other authors declare no competing interests.
Figures



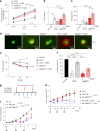

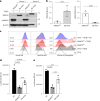

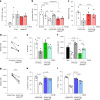


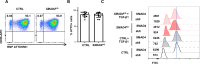

References
-
- Ruggeri, L. et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science295, 2097–2100 (2002). - PubMed
MeSH terms
Substances
Grants and funding
- 765104/EC | EC Seventh Framework Programm | FP7 People: Marie-Curie Actions (FP7-PEOPLE - Specific Programme "People" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- ICI24/00041/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- SGR863/Generalitat de Catalunya (Government of Catalonia)
- 765104/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Sklodowska-Curie Actions (H2020 Excellent Science - Marie Sklodowska-Curie Actions)
- PI21/00002/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- PI22/00040/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- 2024PROD00089/Departament d'Innovació, Universitats i Empresa, Generalitat de Catalunya (Department of Innovation, Education and Enterprise, Government of Catalonia)
- FI23/00075/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- P01 CA111412/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

