Prediction of drug's anatomical therapeutic chemical (ATC) code by constructing biological profiles of ATC codes
- PMID: 40119265
- PMCID: PMC11927162
- DOI: 10.1186/s12859-025-06102-7
Prediction of drug's anatomical therapeutic chemical (ATC) code by constructing biological profiles of ATC codes
Abstract
Background: The Anatomical Therapeutic Chemical (ATC) classification system, proposed and maintained by the World Health Organization, is among the most widely used drug classification schemes. Recently, it has become a key research focus in drug repositioning. Computational models often pair drugs with ATC codes to explore drug-ATC code associations. However, the limited information available for ATC codes constrains these models, leaving significant room for improvement.
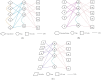
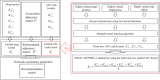
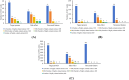
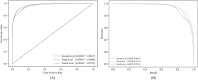
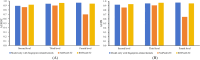
Results: This study presents an inference method to identify highly related target proteins, structural features, and side effects for each ATC code, constructing comprehensive biological profiles. Association networks for target proteins, structural features, and side effects are established, and a random walk with restart algorithm is applied to these networks to extract raw associations. A permutation test is then conducted to exclude false positives, yielding robust biological profiles for ATC codes. These profiles are used to construct new ATC code kernels, which are integrated with ATC code kernels from the existing model PDATC-NCPMKL. The recommendation matrix is subsequently generated using the procedures of PDATC-NCPMKL. Cross-validation results demonstrate that the new model achieves AUROC and AUPR values exceeding 0.96.
Conclusion: The proposed model outperforms PDATC-NCPMKL and other previous models. Analysis of the contributions of the newly added ATC code kernels confirms the value of biological profiles in enhancing the prediction of drug-ATC code associations.
Keywords: Anatomical therapeutic chemical code; Biological profiles; Drug repositioning; Network consistency projection; Random walk with restart.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

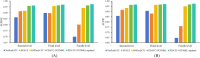
Similar articles
-
PDATC-NCPMKL: Predicting drug's Anatomical Therapeutic Chemical (ATC) codes based on network consistency projection and multiple kernel learning.Comput Biol Med. 2024 Feb;169:107862. doi: 10.1016/j.compbiomed.2023.107862. Epub 2023 Dec 20. Comput Biol Med. 2024. PMID: 38150886
-
Network predicting drug's anatomical therapeutic chemical code.Bioinformatics. 2013 May 15;29(10):1317-24. doi: 10.1093/bioinformatics/btt158. Epub 2013 Apr 5. Bioinformatics. 2013. PMID: 23564845
-
Drug repositioning by prediction of drug's anatomical therapeutic chemical code via network-based inference approaches.Brief Bioinform. 2021 Mar 22;22(2):2058-2072. doi: 10.1093/bib/bbaa027. Brief Bioinform. 2021. PMID: 32221552
-
Convolutional Neural Networks for ATC Classification.Curr Pharm Des. 2018;24(34):4007-4012. doi: 10.2174/1381612824666181112113438. Curr Pharm Des. 2018. PMID: 30417778 Review.
-
Drug-Disease Association Prediction Using Heterogeneous Networks for Computational Drug Repositioning.Biomolecules. 2022 Oct 17;12(10):1497. doi: 10.3390/biom12101497. Biomolecules. 2022. PMID: 36291706 Free PMC article. Review.
Cited by
-
Machine learning approaches reveal methylation signatures associated with pediatric acute myeloid leukemia recurrence.Sci Rep. 2025 May 6;15(1):15815. doi: 10.1038/s41598-025-99258-4. Sci Rep. 2025. PMID: 40328883 Free PMC article.
-
Machine Learning Identifies Key Gene Markers Related to Fetal Retina Development at Single-Cell Transcription Level.Invest Ophthalmol Vis Sci. 2025 Jun 2;66(6):60. doi: 10.1167/iovs.66.6.60. Invest Ophthalmol Vis Sci. 2025. PMID: 40531615 Free PMC article.
-
Unveiling Immune Response Mechanisms in Mpox Infection Through Machine Learning Analysis of Time Series Gene Expression Data.Life (Basel). 2025 Jun 30;15(7):1039. doi: 10.3390/life15071039. Life (Basel). 2025. PMID: 40724541 Free PMC article.
-
Transcriptomic and miRNA Signatures of ChAdOx1 nCoV-19 Vaccine Response Using Machine Learning.Life (Basel). 2025 Jun 18;15(6):981. doi: 10.3390/life15060981. Life (Basel). 2025. PMID: 40566633 Free PMC article.
References
-
- Luo H, Li M, Yang M, Wu FX, Li Y, Wang J. Biomedical data and computational models for drug repositioning: a comprehensive review. Brief Bioinform. 2021;22(2):1604–19. - PubMed
-
- Dowden H, Munro J. Trends in clinical success rates and therapeutic focus. Nat Rev Drug Discov. 2019;18(7):495–6. - PubMed
-
- Nahler G, Nahler G. Anatomical therapeutic chemical classification system (ATC). Dict Pharm Med 2009:8–8.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

