Genomic and proteomic characterization of four novel Schitoviridae family phages targeting uropathogenic Escherichia coli strain
- PMID: 40119445
- PMCID: PMC11927229
- DOI: 10.1186/s12985-025-02691-0
Genomic and proteomic characterization of four novel Schitoviridae family phages targeting uropathogenic Escherichia coli strain
Abstract
Background: Escherichia coli-associated urinary tract infections (UTIs) are among the most prevalent bacterial infections in humans. Typically, antibiotic medication is used to treat UTIs, but over the time, growth of multidrug resistance among these bacteria has created a global public health issue that necessitates other treatment modalities, such as phage therapy.
Methods: The UPEC strain PSU-5266 (UE-17) was isolated from human urine samples, while phages were obtained from wastewater. These phages were characterized through host range analysis, stability studies, adsorption assays, and electron microscopy. Additionally, genomic, phylogenetic, and proteomic analyses were conducted to provide further insights.
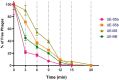
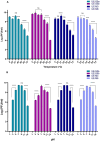
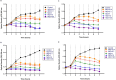
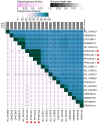
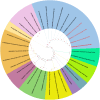
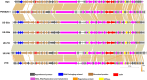
Results: The current study describes the isolation and characterization of four Escherichia coli phages designated as UE-S5a, UE-S5b, UE-M3 and UE-M6. Bactericidal assays depicted that all bacteriophages exhibited a strong lytic ability against uropathogenic E. coli (UPEC) strain PSU-5266 (UE-17). The phages displayed a broad host range (31-41%) among 104 tested isolates and adsorption rate of 15-20 min. They were stable within pH range of 5-11 and temperature range of 4 to 55 °C. Electron microscopy showed that all phages have icosahedral heads (70-74 nm) and short non-contractile tails, thus exhibiting a podovirus morphology. Sequencing results showed that they have linear double stranded DNA, genome of 73 to 76 kb in length, with GC content of 42% and short direct terminal repeats. Their genomes contain 84-88 predicted genes with putative functions predicted to 42-48% of gene products. The phylogenetic and comparative genomic analysis results depicted that these phages, sharing > 98% sequence similarity, are new members of genus Gamaleyavirus of subfamily Enquatrovirinae, in the Schitoviridae family. Mass spectrometric analysis of purified phage particles identified 44-56 phage particle-associated proteins (PPAPs) belonging to various functional groups such as lysis proteins, structural proteins, DNA packaging related proteins, and proteins involved in replication, metabolism and regulation. In addition, no genes encoding virulence factors, antibiotic resistance or lysogeny factors were identified.
Conclusion: The overall findings suggest that these bacteriophages are potential candidates for phage therapy in treating UTIs caused by UPEC strains.
Keywords: Schitoviridae; Phage particle-associated proteins (PPAPs); Phage therapy; Urinary tract infections; Uropathogenic E. coli.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Kline KA, Bowdish DME. Infection in an aging population. Curr Opin Microbiol. 2016;29:63–7. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

