Interleukin expression patterns and immune cell infiltration in prostate adenocarcinoma: Implications for recurrence risk
- PMID: 40119682
- PMCID: PMC11938863
- DOI: 10.1177/03946320251328476
Interleukin expression patterns and immune cell infiltration in prostate adenocarcinoma: Implications for recurrence risk
Abstract
Objective: This study aims to comprehensively investigate the expression profiles of interleukins in prostate adenocarcinoma (PRAD) and their relationship with immune cell infiltration, tumor progression, and patient prognosis. By establishing an interleukin-related risk score, we seek to enhance the understanding of the tumor immune microenvironment and facilitate the development of tailored immunotherapeutic strategies for PRAD patients.
Introduction: Interleukins can nurture a tumor promoting environment and simultaneously regulate immune cell infiltration. However, the potential roles of interleukins in the prostate adenocarcinoma immune landscape remain abstruse.
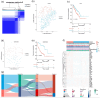
Methods: We comprehensively investigated the interleukin expression patterns and tumor immune landscape of prostate adenocarcinoma patients. And explored the interleukin expression patterns with immune infiltration landscape. The interleukin score was established using LASSO cox regression analysis. Multivariate Cox regression analysis was employed to assess the prognostic value of the interleukin score.
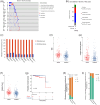
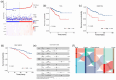
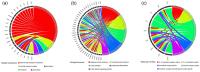
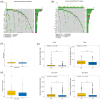
Results: We identified two distinct interleukin clusters, characterized by different immune cell infiltration, tumor promoting signaling pathways activation and prognosis. The interleukin score was established to estimate the prognosis of individual prostate adenocarcinoma (PRAD) patient. Further analysis demonstrated that the interleukin score was an independent prognostic factor of PRAD. Finally, we investigated the predictive value of interleukin score in the programmed cell death protein (PD-1) blockade therapy of patients with prostate adenocarcinoma. At the same time, the differences in related genes among different prostate cell lines were also identified.
Conclusions: This study demonstrated the correlation between interleukin and tumor immune landscape in prostate adenocarcinoma. The comprehensive evaluation of interleukin expression patterns in individual prostate patients contribute to our understanding of the immune landscape and helps clinicians selecting proper immunotherapy strategies for prostate patients.
Keywords: drug selection; immune cell infiltration; immunotherapies; interleukin; prognostic signature; prostate adenocarcinoma.
Conflict of interest statement
Declaration of conflicting interestsThe author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



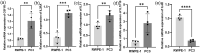
Similar articles
-
Immune infiltration phenotypes of prostate adenocarcinoma and their clinical implications.Cancer Med. 2021 Aug;10(15):5358-5374. doi: 10.1002/cam4.4063. Epub 2021 Jun 15. Cancer Med. 2021. PMID: 34128342 Free PMC article.
-
Single-cell and bulk RNA-sequencing reveals mitosis-involved gene HAUS1 is a promising indicator for predicting prognosis and immune responses in prostate adenocarcinoma (PRAD).Cell Biol Int. 2024 Aug;48(8):1169-1184. doi: 10.1002/cbin.12191. Epub 2024 May 31. Cell Biol Int. 2024. PMID: 38818762
-
Identification of the gene signatures related to NK/T cell communication to evaluate the tumor microenvironment and prognostic outcomes of patients with prostate adenocarcinoma.Front Immunol. 2025 Apr 16;16:1564784. doi: 10.3389/fimmu.2025.1564784. eCollection 2025. Front Immunol. 2025. PMID: 40308606 Free PMC article.
-
Novel immune-related signature for risk stratification and prognosis in prostatic adenocarcinoma.Cancer Sci. 2021 Oct;112(10):4365-4376. doi: 10.1111/cas.15062. Epub 2021 Aug 18. Cancer Sci. 2021. PMID: 34252262 Free PMC article.
-
Roles of m5C RNA Modification Patterns in Biochemical Recurrence and Tumor Microenvironment Characterization of Prostate Adenocarcinoma.Front Immunol. 2022 May 4;13:869759. doi: 10.3389/fimmu.2022.869759. eCollection 2022. Front Immunol. 2022. PMID: 35603206 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

