Medication-resistant epilepsy is associated with a unique gut microbiota signature
- PMID: 40119849
- PMCID: PMC12291004
- DOI: 10.1111/epi.18367
Medication-resistant epilepsy is associated with a unique gut microbiota signature
Abstract
Objective: Dysfunction of the microbiota-gut-brain axis is emerging as a new pathogenic mechanism in epilepsy, potentially impacting on medication response and disease outcome. We investigated the composition of the gut microbiota in a cohort of medication-resistant (MR) and medication-sensitive (MS) pediatric patients with epilepsy.
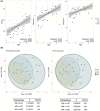
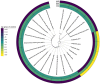
Methods: Children with epilepsy of genetic and presumed genetic etiologies were evaluated clinically and subgrouped into MR and MS. Age-matched healthy controls (HCs) were also recruited. A food diary was used to evaluate nutritional habits, and the Rome IV questionnaire was used to record gastrointestinal symptoms. The microbiota composition was assessed in stool samples through 16S rRNA. α-Diversity (AD) and β-diversity (BD) were calculated, and differential abundance analysis was performed using linear multivariable models (significance: p.adj < .05).
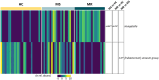
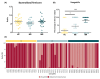
Results: Forty-one patients (MR:MS = 20:21) with a mean age of 7.2 years (±4.6 SD) and 27 age-matched HCs were recruited. No significant differences in AD were found when comparing patients and HCs. Significant positive correlation was found between AD and age (Chao1 p.adj = .0004, Shannon p.adj = .0004, Simpson p.adj = .0028). BD depicted a different bacterial profile in the epilepsy groups compared to HCs (MS vs. HC: Bray-Curtis F = 1.783, p = .001; Jaccard F = 1.24, p = .001; MR vs. HC: Bray-Curtis F = 2.24, p = .001; Jaccard F = 1.364, p = .001). At the genus level, the epilepsy groups were characterized by a significant increase in Hungatella (MS vs. HC: +4.95 log2 change; MR vs. HC: +6.72 log2 change); the [Eubacterium] siraeum group changed between the MR and MS subgroups.
Significance: Epileptic patients display unique gut metagenomic signatures compared to HCs. Moreover, a different ratio of the butyrate-producing [Eubacterium] siraeum group suggests dissimilarities between patients based on the response to antiseizure medications.
Keywords: epilepsy; gut microbiota signature; microbiota–gut–brain axis; pediatrics.
© 2025 The Author(s). Epilepsia published by Wiley Periodicals LLC on behalf of International League Against Epilepsy.
Conflict of interest statement
None of the authors has any conflict of interest to disclose.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

