RNALoc-LM: RNA subcellular localization prediction using pre-trained RNA language model
- PMID: 40119908
- PMCID: PMC11978386
- DOI: 10.1093/bioinformatics/btaf127
RNALoc-LM: RNA subcellular localization prediction using pre-trained RNA language model
Abstract
Motivation: Accurately predicting RNA subcellular localization is crucial for understanding the cellular functions and regulatory mechanisms of RNAs. Although many computational methods have been developed to predict the subcellular localization of lncRNAs, miRNAs, and circRNAs, very few of them are designed to simultaneously predict the subcellular localization of multiple types of RNAs. In addition, the emergence of pre-trained RNA language model has shown remarkable performance in various bioinformatics tasks, such as structure prediction and functional annotation. Despite these advancements, there remains a significant gap in applying pre-trained RNA language models specifically for predicting RNA subcellular localization.
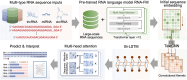
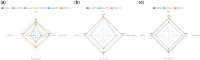
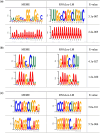
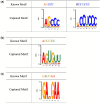
Results: In this study, we proposed RNALoc-LM, the first interpretable deep-learning framework that leverages a pre-trained RNA language model for predicting RNA subcellular localization. RNALoc-LM uses a pre-trained RNA language model to encode RNA sequences, then captures local patterns and long-range dependencies through TextCNN and BiLSTM modules. A multi-head attention mechanism is used to focus on important regions within the RNA sequences. The results demonstrate that RNALoc-LM significantly outperforms both deep-learning baselines and existing state-of-the-art predictors. Additionally, motif analysis highlights RNALoc-LM's potential for discovering important motifs, while an ablation study confirms the effectiveness of the RNA sequence embeddings generated by the pre-trained RNA language model.
Availability and implementation: The RNALoc-LM web server is available at http://csuligroup.com:8000/RNALoc-LM. The source code can be obtained from https://github.com/CSUBioGroup/RNALoc-LM.
© The Author(s) 2025. Published by Oxford University Press.
Figures

References
-
- Ahmad A, Lin H, Shatabda S. Locate-R: subcellular localization of long non-coding RNAs using nucleotide compositions. Genomics 2020;112:2583–9. - PubMed
-
- Asim MN, Ibrahim MA, Zehe C et al. L2S-MirLoc: a lightweight two stage MiRNA sub-cellular localization prediction framework. In: 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China. New York, USA: IEEE, 2021, 1–8.
-
- Bai T, Yan K, Liu B. DAmiRLocGNet: miRNA subcellular localization prediction by combining miRNA–disease associations and graph convolutional networks. Brief Bioinf 2023;24:bbad212. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

