Radial glia integrin avb8 regulates cell autonomous microglial TGFβ1 signaling that is necessary for microglial identity
- PMID: 40121230
- PMCID: PMC11929771
- DOI: 10.1038/s41467-025-57684-y
Radial glia integrin avb8 regulates cell autonomous microglial TGFβ1 signaling that is necessary for microglial identity
Abstract
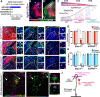
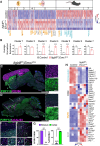
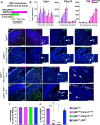
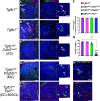
Microglial diversity arises from the interplay between inherent genetic programs and external environmental signals. However, the mechanisms by which these processes develop and interact within the growing brain are not yet fully understood. Here, we show that radial glia-expressed integrin beta 8 (ITGB8) activates microglia-expressed TGFβ1 to drive microglial development. Domain-restricted deletion of Itgb8 in these progenitors results in regionally restricted and developmentally arrested microglia that persist into adulthood. In the absence of autocrine TGFβ1 signaling, microglia adopt a similar phenotype, leading to neuromotor symptoms almost identical to Itgb8 mutant mice. In contrast, microglia lacking the canonical TGFβ signal transducers Smad2 and Smad3 have a less polarized dysmature phenotype and correspondingly less severe neuromotor dysfunction. Our study describes the spatio-temporal regulation of TGFβ activation and signaling in the brain necessary to promote microglial development, and provides evidence for the adoption of microglial developmental signaling pathways in brain injury or disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Drs Butovsky, Arnold and Sheppard have a financial interest in Glial Therapeutics, a company developing a new therapy to target ITGB8- TGFb signaling as a treatment for Alzheimer’s disease. Dr. Butovsky’s interests were reviewed and are managed by BWH and Mass General Brigham in accordance with their conflict of interest policies. The remaining authors declare no competing interests.
Figures

Update of
-
Radial glia promote microglial development through integrin αVβ8 -TGFβ1 signaling.bioRxiv [Preprint]. 2023 Sep 21:2023.07.13.548459. doi: 10.1101/2023.07.13.548459. bioRxiv. 2023. Update in: Nat Commun. 2025 Mar 22;16(1):2840. doi: 10.1038/s41467-025-57684-y. PMID: 37790363 Free PMC article. Updated. Preprint.
References
-
- Squarzoni, P. et al. Microglia modulate wiring of the embryonic forebrain. Cell Rep.8, 1271–1279 (2014). - PubMed
-
- Ueno, M. et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci.16, 543–551 (2013). - PubMed
-
- El Khoury, J. et al. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 13, 432–438 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

