A quantitative analysis of ligand binding at the protein-lipid bilayer interface
- PMID: 40121339
- PMCID: PMC11929912
- DOI: 10.1038/s42004-025-01472-8
A quantitative analysis of ligand binding at the protein-lipid bilayer interface
Abstract
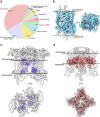
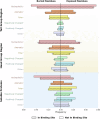
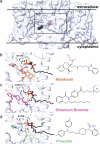
The majority of drugs target membrane proteins, and many of these proteins contain ligand binding sites embedded within the lipid bilayer. However, targeting these therapeutically relevant sites is hindered by limited characterization of both the sites and the molecules that bind to them. Here, we introduce the Lipid-Interacting LigAnd Complexes Database (LILAC-DB), a curated dataset of 413 structures of ligands bound at the protein-bilayer interface. Analysis of these structures reveals that ligands binding to lipid-exposed sites exhibit distinct chemical properties, such as higher calculated partition coefficient (clogP), molecular weight, and a greater number of halogen atoms, compared to ligands that bind to soluble proteins. Additionally, we demonstrate that the atomic properties of these ligands vary significantly depending on their depth within and exposure to the lipid bilayer. We also find that ligand binding sites exposed to the bilayer have distinct amino acid compositions compared to other protein regions, which may aid in the identification of lipid-exposed binding sites. This analysis provides valuable guidelines for researchers pursuing structure-based drug discovery targeting underexploited ligand binding sites at the protein-lipid bilayer interface.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Payandeh, J. & Volgraf, M. Ligand binding at the protein–lipid interface: strategic considerations for drug design. Nat. Rev. Drug Discov.20, 710–722 (2021). - PubMed
-
- Wang, Y., Yu, Z., Xiao, W., Lu, S. & Zhang, J. Allosteric binding sites at the receptor–lipid bilayer interface: novel targets for GPCR drug discovery. Drug Discov. Today26, 690–703 (2021). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

