Immunoglobulins G from Patients with Systemic Sclerosis Modify the Molecular Signatures of Endothelial Cells
- PMID: 40122572
- PMCID: PMC11931898
- DOI: 10.1136/rmdopen-2024-004290
Immunoglobulins G from Patients with Systemic Sclerosis Modify the Molecular Signatures of Endothelial Cells
Abstract
Objective: Antinuclear antibodies (ANA) are powerful biomarkers in systemic sclerosis (SSc). Functional antibodies (FA) might be implicated in vasculopathy, in which endothelial cells (EC) are key players. We aimed to explore the effect of purified IgG from patients with SSc on omics signatures of EC and examine the influence of ANA serotypes and FA.
Methods: EC were cultured in the presence of purified IgG from patients with SSc, patients with systemic lupus erythematosus (SLE) or healthy controls (HC). EC omics profiles were analysed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) and RNA sequencing. EC proteome induced by IgG from patients with SSc was confirmed with an external validation cohort.
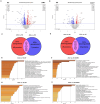
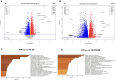
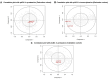
Results: In the derivation cohort, principal component analysis (PCA) using proteomics data showed three distinct groups of subjects: a first one including mostly anti-topoisomerase-I positive patients (ATA+), a second one including mostly anti-centromere positive patients and a third group comprising anti-RNA polymerase-III positive patients, SLE and HC. In transcriptomics, PCA distinguished one group composed of ATA+patients only from a second group mixing ATA+patients with other individuals. The validation cohort confirmed the existence of two groups of distinct EC proteome profiles and clinical severity in ATA+patients. In both SSc cohorts, no association between FA presence and proteomic profiles was observed. Quantitative proteomics measured the most discriminant proteins in EC exposed to purified IgG.
Conclusion: Purified IgG from patients with SSc can modify EC proteome and transcriptome. The observed changes closely associate with ANA serotype.
Keywords: Autoantibodies; Autoimmune Diseases; Scleroderma, Systemic.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: AC reports a relationship with Vitalair that includes: travel reimbursement. DL reports consultancies and speaking fees from Biocryst Pharmaceuticals, Inc., CSL Behring LLC, Takeda Pharmaceutical Company Limited and research support from Biocryst Pharmaceuticals Inc., CSL Behring LLC, GSK, Octapharma, Pfizer Inc. and Takeda Pharmaceutical Company Limited (less than US$10 000), outside the submitted work. VS reports consultancies and speaking fees from Boehringer-Ingelheim, Fresenius Kabi, Grifols, Ultragenyx (less than US$10 000) and research support from Grifols, outside the submitted work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
