Single-cell sequencing combined with urinary multi-omics analysis reveals that the non-invasive biomarker PRDX5 regulates bladder cancer progression through ferroptosis signaling
- PMID: 40122834
- PMCID: PMC11931876
- DOI: 10.1186/s12885-025-13881-y
Single-cell sequencing combined with urinary multi-omics analysis reveals that the non-invasive biomarker PRDX5 regulates bladder cancer progression through ferroptosis signaling
Abstract
Objective: This study aimed to elucidate the expression profile and biological implications of peroxidase 5 (PRDX5) in bladder cancer (BC), specifically investigating its influence on BC progression through modulation of reactive oxygen species (ROS) levels and activation of ferroptosis pathways.
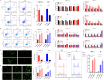
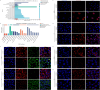
Methods: We employed urine proteomics data and transcriptomic information from the Cancer Genome Atlas (TCGA) to identify differentially expressed genes in BC tissues, focusing on PRDX5. Using single-cell RNA sequencing (scRNA-seq), we assessed PRDX5 distribution across various cell types in the tumor microenvironment. We conducted in vitro experiments to analyze the impact of PRDX5 on BC cell proliferation, migration, and invasion, while exploring its mechanisms of modulating ROS levels and ferroptosis. In vivo experiments were performed to observe PRDX5's influence on ferroptosis signaling in tissue contexts.
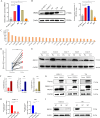
Results: We found significant upregulation of PRDX5 in BC tissues, with scRNA-seq revealing its enrichment in bladder epithelial cells, correlating with disease advancement and established BC markers. In vitro analyses showed that overexpressed PRDX5 enhanced proliferation, migration, and invasion of BC cells, while PRDX5 knockout produced opposing effects. Additionally, PRDX5 modulated ROS levels and impacted ferroptosis pathways. In vivo experiments confirmed that PRDX5 knockout inhibited tumor growth and activated ferroptosis signaling pathways in tissues.
Conclusion: Our study highlights the elevated expression of PRDX5 in BC and its role in promoting tumor progression through regulation of ROS levels and ferroptosis. PRDX5 may serve as a promising target for BC treatment, supporting further exploration of its potential in clinical applications.
Keywords: Bladder cancer; Ferroptosis; Multi-omics; PRDX5; Progression; Single cell; Urine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Our study adhered to the Declaration of Helsinki and ethical guidelines established by the Institutional Medical Ethics Committee of Lanzhou University Second Hospital. All clinical samples were collected with informed consent from the patients and the collection was approved by the Medical Ethics Committee of the Lanzhou University Second Hospital (D2024-774). Consent for publication: Not applicable. Competing Interest: The authors declare no competing interests.
Figures







Similar articles
-
Characterization of the Biochemical Recurrence Prediction Ability and Progression Correlation of Peroxiredoxins Family in Prostate Cancer Based on Integrating Single-Cell RNA-Seq and Bulk RNA-Seq Cohorts.Cancer Med. 2025 May;14(9):e70855. doi: 10.1002/cam4.70855. Cancer Med. 2025. PMID: 40281661 Free PMC article.
-
Squalene epoxidase promotes breast cancer progression by regulating CCNB1 protein stability.Exp Cell Res. 2023 Dec 1;433(1):113805. doi: 10.1016/j.yexcr.2023.113805. Epub 2023 Oct 14. Exp Cell Res. 2023. PMID: 37839786
-
Integrating single-cell transcriptomics to reveal the ferroptosis regulators in the tumor microenvironment that contribute to bladder urothelial carcinoma progression and immunotherapy.Front Immunol. 2024 Aug 22;15:1427124. doi: 10.3389/fimmu.2024.1427124. eCollection 2024. Front Immunol. 2024. PMID: 39238647 Free PMC article.
-
Involvement of the PRDX family and its clinical functions in different types of gastrointestinal cancer (Review).Oncol Rep. 2025 Aug;54(2):90. doi: 10.3892/or.2025.8923. Epub 2025 Jun 6. Oncol Rep. 2025. PMID: 40476576 Review.
-
Sex differences in bladder cancer: understanding biological and clinical implications.Biol Sex Differ. 2025 May 13;16(1):31. doi: 10.1186/s13293-025-00715-6. Biol Sex Differ. 2025. PMID: 40361239 Free PMC article. Review.
References
-
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12–49. - PubMed
-
- Eissa S, Swellam M, el-Mosallamy H, Mourad MS, Hamdy NΜ, Kamel K, et al. Diagnostic value of urinary molecular markers in bladder cancer. Anticancer Res. 2003;23(5b):4347–55. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Pakmanesh H, Khajehsalimi A, Hesamarefi M, Ebadzadeh MR, Bazrafshan A, Malekpourafshar R, et al. Comparison of the overall survival of different treatment methods in patients with muscle-invasive bladder cancer: a retrospective study. Urologia. 2024;91(4):687–94. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

