Proximity proteomics reveals unique and shared pathological features between multiple system atrophy and Parkinson's disease
- PMID: 40122840
- PMCID: PMC11931798
- DOI: 10.1186/s40478-025-01958-5
Proximity proteomics reveals unique and shared pathological features between multiple system atrophy and Parkinson's disease
Abstract
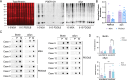
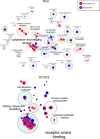
Synucleinopathies such as Parkinson's disease (PD), dementia with Lewy bodies (DLB), and multiple system atrophy (MSA) are neurodegenerative diseases with shared clinical and pathological features. Aggregates of alpha-synuclein (αsyn) phosphorylated at serine 129 (PSER129) are hallmarks of synucleinopathies, which, for PD/DLB, are found predominantly in neurons, whereas in MSA, aggregates are primarily found in oligodendroglia. It remains unclear whether the distinct pathological presentations of PD/DLB and MSA are manifestations of unique or shared pathological processes. Using the in-situ proximity labeling technique of biotinylation by antibody recognition (BAR), we compared aggregated αsyn-interactomes (BAR-PSER129) and total αsyn-interactomes (BAR-MJFR1) between MSA (n = 5) and PD/DLB (n = 10) in forebrain and midbrain structures. Comparison between MSA and PD/DLB-enriched proteins revealed 79 PD/DLB-differentially abundant proteins and only three MSA-differentially abundant proteins (CBR1, CRYAB, and GFAP). Pathway enrichment analysis revealed that vesicle/SNARE-associated pathways dominated PD/DLB interactions, whereas MSA was strongly enriched for metabolic/catabolic, iron, and cellular oxidant detoxification pathways. A subnetwork of cytosolic antioxidant enzymes called peroxiredoxins drove cellular detoxification pathway enrichment in MSA. A network of 26 proteins, including neuronal-specific proteins (e.g., SYNGR3) with HSPA8 at the core, was shared between MSA and DLB/PD. Extracellular exosome pathways were universally enriched regardless of the disease or BAR target protein. In conclusion, synucleinopathies have divergent and convergent αsyn-aggregate interactions, indicating unique and shared pathogenic mechanisms. MSA uniquely involves oxidant detoxification processes in glial cells, while vesicular processes in neurons dominate PD/DLB. Shared interactions, specifically SYNGR3, between MSA and PD/DLB suggest that neuronal axons are the origin of both diseases. In conclusion, we provide αsyn protein interaction maps for two distinct synucleinopathies.
Keywords: Antioxidant; Neurodegeneration; Proximity proteomics; SNARE complex; Spatial omics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Brain specimens were provided by the Rush Movement Disorders Brain Bank, which collects high quality brain specimens with Rush University institutional review board approval. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Update of
-
Comparing alpha-synuclein-interactomes between multiple systems atrophy and Parkinson's disease reveals unique and shared pathological features.bioRxiv [Preprint]. 2024 Sep 21:2024.09.20.613717. doi: 10.1101/2024.09.20.613717. bioRxiv. 2024. Update in: Acta Neuropathol Commun. 2025 Mar 23;13(1):65. doi: 10.1186/s40478-025-01958-5. PMID: 39345456 Free PMC article. Updated. Preprint.
References
-
- Bjornsdottir A, Gudmundsson G, Blondal H, Olafsson E (2013) Incidence and prevalence of multiple system atrophy: a nationwide study in Iceland. J Neurol Neurosurg Psychiatry 84:136–140. 10.1136/jnnp-2012-302500 - PubMed
-
- Borghi R, Marchese R, Negro A, Marinelli L, Forloni G, Zaccheo D, Abbruzzese G, Tabaton M (2000) Full length alpha-synuclein is present in cerebrospinal fluid from Parkinson’s disease and normal subjects. Neurosci Lett 287:65–67. 10.1016/s0304-3940(00)01153-8 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

