A bioinformatics approach combined with experimental validation analyzes the efficacy of azithromycin in treating SARS-CoV-2 infection in patients with IPF and COPD These authors contributed equally: Yining Xie, Guangshu Chen, and Weiling Wu
- PMID: 40122903
- PMCID: PMC11930991
- DOI: 10.1038/s41598-025-94801-9
A bioinformatics approach combined with experimental validation analyzes the efficacy of azithromycin in treating SARS-CoV-2 infection in patients with IPF and COPD These authors contributed equally: Yining Xie, Guangshu Chen, and Weiling Wu
Abstract
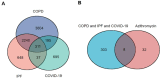
The swift transmission rate and unfavorable prognosis associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have prompted the pursuit of more effective therapeutic interventions. Azithromycin (AZM) has garnered significant attention for its distinctive pharmacological mechanisms in the treatment of SARS-CoV-2. This study aims to elucidate the biological rationale for employing AZM in patients with chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF) who are infected with SARS-CoV-2. Genetic data about COVID-19, COPD, and IPF were independently obtained from the GeneCards database. And 40 drug targets about AZM were retrieved from the STITCH database. The analysis revealed that 311 DEGs were common among COPD, IPF, and COVID-19, and we further found eight genes that interacted with AZM targets. We conducted an analysis of hub genes and their corresponding signaling pathways in these patient cohorts. Additionally, we explored the inhibitory effects of AZM on these hub genes. AZM demonstrated a significant inhibitory effect on eight key genes, except for AR and IL-17 A. These findings suggest that AZM may serve as a promising therapeutic agent for patients with COPD and IPF and SARS-CoV-2 infection.
Keywords: Azithromycin; Bioinformatics; COPD; IPF; SARS-Cov-2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable.
Figures



Similar articles
-
Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease patients.Brief Bioinform. 2021 Sep 2;22(5):bbab115. doi: 10.1093/bib/bbab115. Brief Bioinform. 2021. PMID: 33847347 Free PMC article.
-
Network-based identification genetic effect of SARS-CoV-2 infections to Idiopathic pulmonary fibrosis (IPF) patients.Brief Bioinform. 2021 Mar 22;22(2):1254-1266. doi: 10.1093/bib/bbaa235. Brief Bioinform. 2021. PMID: 33024988 Free PMC article.
-
Identification of hub genes associated with COVID-19 and idiopathic pulmonary fibrosis by integrated bioinformatics analysis.PLoS One. 2022 Jan 19;17(1):e0262737. doi: 10.1371/journal.pone.0262737. eCollection 2022. PLoS One. 2022. PMID: 35045126 Free PMC article.
-
Azithromycin: Immunomodulatory and antiviral properties for SARS-CoV-2 infection.Eur J Pharmacol. 2021 Aug 15;905:174191. doi: 10.1016/j.ejphar.2021.174191. Epub 2021 May 17. Eur J Pharmacol. 2021. PMID: 34015317 Free PMC article. Review.
-
Azithromycin in viral infections.Rev Med Virol. 2021 Mar;31(2):e2163. doi: 10.1002/rmv.2163. Epub 2020 Sep 23. Rev Med Virol. 2021. PMID: 32969125 Free PMC article. Review.
References
-
- CDC. COVID Data Tracker. Variant Proportions. https://covid.cdc.gov/covid-data-tracker/#variant-proportions
-
- Organization, W. H. Therapeutics and COVID-19: Living guideline, 10 November (2023). https://www.who.int/publications/i/item/WHO-2019-nCoV-therapeutics-2023.2
-
- Lee, S. H. et al. Clinical features and prognosis of patients with idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease. Int. J. Tuberc Lung Dis.23, 678–684. 10.5588/ijtld.18.0194 (2019). - PubMed
-
- Halpin, D. & Prevention of Chronic Obstructive Lung Disease. Global Initiative for the Diagnosis, Management, and. The 2020 GOLD Science Committee Report on COVID-19 and Chronic Obstructive Pulmonary Disease. American journal of respiratory and critical care medicine 203, 24–36, (2021). 10.1164/rccm.202009-3533SO - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

