Evaluation of the Diagnostic Accuracy of the T2Resistance Panel (Research Use Only) in Patients With Possible Bacterial Bloodstream Infections
- PMID: 40123928
- PMCID: PMC11924974
- DOI: 10.31486/toj.24.0101
Evaluation of the Diagnostic Accuracy of the T2Resistance Panel (Research Use Only) in Patients With Possible Bacterial Bloodstream Infections
Erratum in
-
ERRATUM.Ochsner J. 2025 Summer;25(2):74. doi: 10.31486/toj.25.5054. Ochsner J. 2025. PMID: 40538603 Free PMC article.
Abstract
Background: Early identification and antimicrobial susceptibility testing (AST) of bloodstream pathogens are important for promptly determining the appropriate therapy. Currently, positive blood culture results (identification and AST) are reported in 2 to 4 days. The T2Resistance (T2R) Panel (T2 Biosystems, Inc) uses DNA amplification with magnetic resonance from 3 mL of whole blood for direct detection of 13 antibiotic resistance genes: bla KPC, bla NDM, bla VIM, bla IMP, bla OXA-48, bla CTX-M-14/15, AmpC bla CMY/DHA, vanA/B, and mecA/C. We compared the accuracy of T2R testing to AST for positive blood cultures.
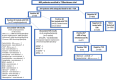
Methods: This investigator-sponsored, single-center study prospectively enrolled 802 patients with a standard of care blood culture. Five hundred forty-seven patients had adequate blood for culture and T2R testing. Blood cultures with positive isolates were identified, and AST was performed. Blood samples with positive blood cultures were tested with the T2R Panel.
Results: Blood cultures were positive for 58/547 (10.6%) patients. Contaminants (18/547 [3.3%]) were excluded. T2R testing results (n=31) showed 2 bla CTX-M-14/15 genes with 100% sensitivity; the remaining gram-negative resistance genes were not detected, so sensitivity could not accurately be determined. Specificity was 100% for the 16 gram-negative bacilli. Three enterococci and 2 Staphylococcus aureus showed 100% sensitivity/specificity. However, 10 coagulase-negative staphylococci showed 17% sensitivity/100% specificity. Antibiotic resistance genes identified were 2 bla CTX-M-14/15, 2 mecA/C, and 1 vanA/B. T2R testing results were obtained in an average of 7 hours.
Conclusion: T2R testing is highly specific (100%) for the 13 antibiotic resistance genes on the panel. Sensitivity was 100% for the genes detected but was low (17%) for coagulase-negative staphylococci. T2R testing has the potential to diagnose certain antibiotic resistance genes from bacterial bloodstream infections in hours vs the days required for a positive blood culture with AST. Additional studies that include larger numbers of samples with antimicrobial resistance genes are needed.
Keywords: Drug resistance–microbial; genes–MDR; rapid diagnostic tests.
©2025 by the author(s); Creative Commons Attribution License (CC BY).
Conflict of interest statement
Portions of these data were presented at the ASM Microbe meeting, Washington, DC, June 12, 2022. The authors thank T2 Biosystems for providing the T2Resistance Panels (research use only) for this investigator-initiated study and Azenta, Inc for performing the next-generation sequencing and analysis on the 9 bacterial isolates from blood cultures that tested negative with the T2R Panel. The authors have no financial or proprietary interest in the subject matter of this article.
Figures
References
-
- World Health Organization. Global Report on the Epidemiology and Burden of Sepsis: Current Evidence, Identifying Gaps and Future Directions. Published September 9, 2020. Accessed March 2, 2025. who.int/publications/i/item/9789240010789
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
