Construction and validation of machine learning-based predictive model for colorectal polyp recurrence one year after endoscopic mucosal resection
- PMID: 40124266
- PMCID: PMC11924002
- DOI: 10.3748/wjg.v31.i11.102387
Construction and validation of machine learning-based predictive model for colorectal polyp recurrence one year after endoscopic mucosal resection
Abstract
Background: Colorectal polyps are precancerous diseases of colorectal cancer. Early detection and resection of colorectal polyps can effectively reduce the mortality of colorectal cancer. Endoscopic mucosal resection (EMR) is a common polypectomy procedure in clinical practice, but it has a high postoperative recurrence rate. Currently, there is no predictive model for the recurrence of colorectal polyps after EMR.
Aim: To construct and validate a machine learning (ML) model for predicting the risk of colorectal polyp recurrence one year after EMR.
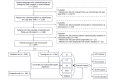
Methods: This study retrospectively collected data from 1694 patients at three medical centers in Xuzhou. Additionally, a total of 166 patients were collected to form a prospective validation set. Feature variable screening was conducted using univariate and multivariate logistic regression analyses, and five ML algorithms were used to construct the predictive models. The optimal models were evaluated based on different performance metrics. Decision curve analysis (DCA) and SHapley Additive exPlanation (SHAP) analysis were performed to assess clinical applicability and predictor importance.
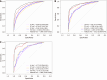
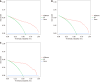
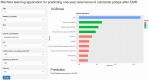
Results: Multivariate logistic regression analysis identified 8 independent risk factors for colorectal polyp recurrence one year after EMR (P < 0.05). Among the models, eXtreme Gradient Boosting (XGBoost) demonstrated the highest area under the curve (AUC) in the training set, internal validation set, and prospective validation set, with AUCs of 0.909 (95%CI: 0.89-0.92), 0.921 (95%CI: 0.90-0.94), and 0.963 (95%CI: 0.94-0.99), respectively. DCA indicated favorable clinical utility for the XGBoost model. SHAP analysis identified smoking history, family history, and age as the top three most important predictors in the model.
Conclusion: The XGBoost model has the best predictive performance and can assist clinicians in providing individualized colonoscopy follow-up recommendations.
Keywords: Colorectal polyps; Machine learning; Predictive model; Risk factors; SHapley Additive exPlanation.
©The Author(s) 2025. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Figures

References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. - PubMed
-
- Dekker E, Rex DK. Advances in CRC Prevention: Screening and Surveillance. Gastroenterology. 2018;154:1970–1984. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

