Genomic characteristics of emerging human pathogen Rickettsia aeschlimannii isolated from two Hyalomma tick species
- PMID: 40124488
- PMCID: PMC11930227
- DOI: 10.1016/j.isci.2025.112080
Genomic characteristics of emerging human pathogen Rickettsia aeschlimannii isolated from two Hyalomma tick species
Abstract
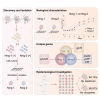
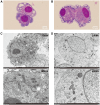
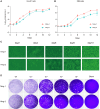
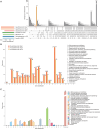
Rickettsia aeschlimannii, which emerged in Morocco in 1997, causes the Mediterranean spotted fever-like rickettsiosis in various Mediterranean countries and recently in Russia and China. Despite its increasing distribution, no available genome has been reported outside Morocco to date. Here, we isolated two strains of R. aeschlimannii from Hyalomma asiaticum (Ning-1 strain) and Hyalomma scupense (Ning-2 strain) ticks in northwestern China and assembled their complete genomes. The genomes of the two strains were smaller than the Mediterranean MC16 strain, containing fewer pseudogenes, higher ralF virulence factor coverage, and 154 unique orthogroups. The Ning-1 strain overwhelmed the Ning-2 strain with more obvious cytopathic effects, quicker growth, and faster plaque formation in cell culture, likely due to its unique pmp20 gene, higher frequency of single nucleotide polymorphisms, and missense/silent ratio. The prevalence of R. aeschlimannii was high among Hyalomma ticks in northwestern China. These findings highlight the genomic characteristics of R. aeschlimannii and the necessity for enhanced surveillance of the emerging Rickettsia in the human population.
Keywords: Genomics; Pathogenic organism.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Rickettsia aeschlimannii Infection in a Woman from Xingjiang, Northwestern China.Vector Borne Zoonotic Dis. 2022 Jan;22(1):55-57. doi: 10.1089/vbz.2021.0071. Vector Borne Zoonotic Dis. 2022. PMID: 35030047
-
First report of spotted fever group Rickettsia aeschlimannii in Hyalomma turanicum, Haemaphysalis bispinosa, and Haemaphysalis montgomeryi infesting domestic animals: updates on the epidemiology of tick-borne Rickettsia aeschlimannii.Front Microbiol. 2023 Dec 15;14:1283814. doi: 10.3389/fmicb.2023.1283814. eCollection 2023. Front Microbiol. 2023. PMID: 38163073 Free PMC article.
-
Molecular detection of Rickettsia aeschlimannii, Candidatus Rickettsia shennongii, Rickettsia sp. and Coxiella burnetii in ticks collected from camels.Sci Rep. 2024 Sep 27;14(1):22129. doi: 10.1038/s41598-024-73663-7. Sci Rep. 2024. PMID: 39333333 Free PMC article.
-
Draft Genome Sequence of Rickettsia aeschlimannii, Associated with Hyalomma marginatum Ticks.Genome Announc. 2014 Jul 24;2(4):e00666-14. doi: 10.1128/genomeA.00666-14. Genome Announc. 2014. PMID: 25059861 Free PMC article.
-
Tick-borne rickettsioses in Europe.Ticks Tick Borne Dis. 2012 Dec;3(5-6):271-8. doi: 10.1016/j.ttbdis.2012.10.035. Epub 2012 Nov 21. Ticks Tick Borne Dis. 2012. PMID: 23177355 Review.
Cited by
-
Serological and molecular insights into tick-borne pathogens in wild donkeys from an unexplored Mediterranean nature reserve.Curr Res Parasitol Vector Borne Dis. 2025 May 7;7:100267. doi: 10.1016/j.crpvbd.2025.100267. eCollection 2025. Curr Res Parasitol Vector Borne Dis. 2025. PMID: 40503032 Free PMC article.
References
LinkOut - more resources
Full Text Sources

