Evolving concepts of the protein universe
- PMID: 40124498
- PMCID: PMC11926713
- DOI: 10.1016/j.isci.2025.112012
Evolving concepts of the protein universe
Abstract
The protein universe is the collection of all proteins on earth from all organisms both extant and extinct. Classical studies on protein folding suggested that proteins exist as a unique three-dimensional conformation that is dictated by the genetic code and is critical for function. In this perspective, we discuss ideas and developments that emerged over the past three decades regarding the protein structure-function paradigm. It is now clear that ordered (active/functional) and disordered/denatured (and hence inactive/non-functional) represent a continuum of states rather than binary states. Some proteins can switch folds without sequence change. Others exist as conformational ensembles lacking defined structure yet play critical roles in many biological processes, including forming membrane-less organelles driven by liquid-liquid phase separation. Numerous diverse proteins harbor segments with the potential to form amyloid fibrils, many of which are functional, and some possess prion-like properties enabling conformation-based transfer of heritable information. Taken together, these developments reveal the remarkable complexity of the protein universe.
Keywords: Biochemistry; Biological sciences; Protein; Structural biology.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




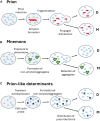

References
-
- Ladunga I. Phylogenetic continuum indicates "galaxies" in the protein universe: preliminary results on the natural group structures of proteins. J. Mol. Evol. 1992;34:358–375. - PubMed
-
- Povolotskaya I.S., Kondrashov F.A. Sequence space and the ongoing expansion of the protein universe. Nature. 2010;465:922–926. - PubMed
-
- Matveev V.V. Cell theory, intrinsically disordered proteins, and the physics of the origin of life. Prog. Biophys. Mol. Biol. 2019;149:114–130. - PubMed
-
- Kauzmann W.J. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959;14:1–63. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

