Methionine Metabolism Dictates PCSK9 Expression and Antitumor Potency of PD-1 Blockade in MSS Colorectal Cancer
- PMID: 40125618
- PMCID: PMC12097065
- DOI: 10.1002/advs.202501623
Methionine Metabolism Dictates PCSK9 Expression and Antitumor Potency of PD-1 Blockade in MSS Colorectal Cancer
Abstract
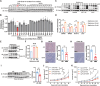
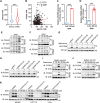
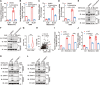
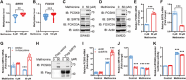
Nutrient metabolisms are vitally interrelated to cancer progression and immunotherapy. However, the mechanisms by which nutrient metabolisms interact to remodel immune surveillance within the tumor microenvironment remain largely unexplored. Here it is demonstrated that methionine restriction inhibits the expression of proprotein convertase subtilisin/kexin type 9 (PCSK9), a key regulator of cholesterol homeostasis and a potential target for cancer immunotherapy, in colorectal cancer (CRC) but not in the liver. Mechanistically, methionine is catabolized to S-adenosylmethionine (SAM), promoting mRNA transcription of PCSK9 through increased DNA methyltransferase 1 (DNMT1)-mediated DNA methylation and suppression of sirtuin 6 (SIRT6) expression. Furthermore, both PCSK9 inhibition and dietary methionine restriction (DMR) potentiate PD-1 blockade therapy and foster the infiltration of CD8+ T cells in Colon 26 tumor-bearing mice-a proficient mismatch repair (pMMR)/microsatellite stable (MSS) CRC model that exhibits limited response to anti-PD-1 therapy. Moreover, combining 5-fluorouracil (5-FU) chemotherapy with PCSK9 inhibition and PD-1 blockade further augments therapeutic efficacy for MSS CRC. The findings establish a mechanistic link between amino acid metabolism and cholesterol metabolism within the tumor microenvironment where tumor cells sense methionine to regulate PCSK9 expression, highlighting promising combination therapeutic strategies that may greatly benefit MSS CRC patients.
Keywords: MSS colorectal cancer; PCSK9; immunotherapy resistance; methionine metabolism.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wu J., Li G., Li L., Li D., Dong Z., Jiang P., Nat. Cell Biol. 2021, 23, 75. - PubMed
MeSH terms
Substances
Grants and funding
- 2018YFA0800404/National Key R&D Program of China
- 82372774/National Natural Science Foundation of China
- 81822036/National Natural Science Foundation of China
- 31770931/National Natural Science Foundation of China
- 31800730/National Natural Science Foundation of China
- 82001745/National Natural Science Foundation of China
- 2023B1111020008/Key-Area Research and Development Program of Guangdong Province
- 2017A030306030/Guangdong Natural Science Funds for Distinguished Young Scholar
- 2017GC010332/Pearl River Talent Plan of Guangdong Province
- 2018030310293/Natural Science Foundation of Guangdong Province
- 2024A1515010664/Guangdong Basic and Applied Basic Research Foundation
- 2020A1515011246/Guangdong Basic and Applied Basic Research Foundation
- 2019A1515110052/Guangdong Basic and Applied Basic Research Foundation
- 202201011490/Guangzhou Science and Technology Plan Project
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
