Development of MDM2-Targeting PROTAC for Advancing Bone Regeneration
- PMID: 40125646
- PMCID: PMC12097015
- DOI: 10.1002/advs.202415626
Development of MDM2-Targeting PROTAC for Advancing Bone Regeneration
Abstract
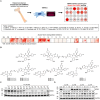
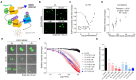
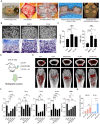
Proteolysis-targeting chimeras (PROTACs) degrade target proteins through the ubiquitin-proteasome system. To date, PROTACs are primarily used to treat various diseases; however, they have not been applied in regenerative therapy. Herein, this work introduces MDM2-targeting PROTACs customized for application in bone regeneration. An MDM2-PROTAC library is constructed by combining Nutlin-3 and CRBN ligands with various linker designs. Through a multistep validation process, this work develops MDM2-PROTACs (CL144 and CL174) that presented potent degradation efficiency and a robust inductive effect on the biomineralization. Next, this work performs whole-transcriptome analysis to dissect the biological effects of the CL144, and reveals the upregulation of osteogenic marker genes. Furthermore, CL144 effectively induced bone regeneration in bone graft and ovariectomy (OVX) models after local and systemic administration, respectively. In the OVX model, the combination treatment with CL144 and alendronate induced a synergistic effect. Overall, this study demonstrates the promising role of MDM2-PROTAC in promoting bone regeneration, marking the first step toward expanding the application of the PROTAC technology.
Keywords: MDM2; PROTAC; bone; osteoporosis; regenerative medicine.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Soliman H., Theret M., Scott W., Hill L., Underhill T. M., Hinz B., Rossi F. M. V., Cell Stem Cell 2021, 28, 1690. - PubMed
-
- a) James A. W., LaChaud G., Shen J., Asatrian G., Nguyen V., Zhang X., Ting K., Soo C., Tissue Eng. Part B 2016, 22, 284; - PMC - PubMed
- b) Pan H. C., Lee S., Ting K., Shen J., Wang C., Nguyen A., Berthiaume E. A., Zara J. N., Turner A. S., H. B. Seim 3rd, Kwak J. H., Zhang X., Soo C., Am. J. Pathol. 2017, 187, 1485. - PMC - PubMed
-
- Nakashima M., Reddi A. H., Nat. Biotechnol. 2003, 21, 1025. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
