A novel Erwiniaceae gut symbiont modulates gene expression of the intracellular bacterium Cardinium in the stored product mite Tyrophagus putrescentiae
- PMID: 40126013
- PMCID: PMC12039267
- DOI: 10.1128/msphere.00879-24
A novel Erwiniaceae gut symbiont modulates gene expression of the intracellular bacterium Cardinium in the stored product mite Tyrophagus putrescentiae
Abstract
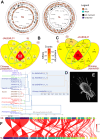
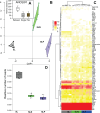
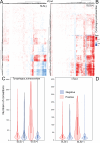
We examined host and bacterial gene expression profiles in the stored product mite Tyrophagus putrescentiae co-infected with Wolbachia (wTPut) and Cardinium (cTPut) while varying the presence of the Erwiniaceae symbiont (SLS). SLS, a novel symbiont in the family Erwiniaceae, with a genome size of 1.7 Mb, is found in 16% of mite species in infected cultures. In addition, SLS was detected in mite feces but not in their eggs. Although Wolbachia expression remained unchanged, the presence or absence of SLS significantly affected Cardinium expression. It indicated that the effect of Wolbachia on SLS was neutral. In SLS-positive samples, Cardinium exhibited 29 upregulated and 48 downregulated genes compared to SLS-negative samples. Furthermore, Cardinium gene expression strongly correlated with mite KEGG gene expression in SLS-positive samples. Positive Spearman's correlations between Cardinium gene expression and mite KEGG immune and regulatory pathways were doubled in SLS-positive compared to SLS-negative samples. The diversity of expressed genes in the mite host decreased in the presence of SLS. Cardinium had more interacting genes to mite host in SLS-positive samples than without SLS. Transposases are the most affected Cardinium genes, showing upregulation in the presence of SLS. Correlation analyses revealed interactions between Cardinium and SLS via mite immune and regulatory pathways, including lysosome, ubiquitin-mediated proteolysis, PIK3_Akt, and cGMP-PKG. The results showed that Cardinium indirectly affects the gut symbionts of mites.IMPORTANCEThis study introduces a new model to analyze interactions between intracellular bacterial symbionts, gut bacterial symbionts, and their mite hosts. Using gene expression correlations, we investigated how the intracellular Cardinium responds to the novel Erwiniaceae gut symbiont in the mold mite Tyrophagus putrescentiae. The data showed that both mite and Cardinium gene expression are different in the samples with and without Erwiniaceae symbionts. In the presence of Erwiniaceae symbionts, Cardinium increased the interaction with the mite host in terms of changes in gene expression. The mite immune and regulatory pathway gene expression is differently correlated to Cardinium genes in relation to Erwiniaceae symbionts. As a well-known producer of allergens, T. putrescentiae physiology and thus its allergen production are influenced by both symbionts, potentially affecting the release of allergens into human environments.
Keywords: Cardinium; Erwiniaceae; Sodalis; Tyrophagus putrescentiae; Wolbachia; allergens; bacterial symbionts; gene expression; stored product mite.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Analysis of the tripartite interactions between two bacterial symbionts, a novel Solitalea-like bacterium (Bacteroidota) and Cardinium, and the stored product mite Tyrophagus putrescentiae based on gene expression data.Microbiol Spectr. 2025 Aug 5;13(8):e0060925. doi: 10.1128/spectrum.00609-25. Epub 2025 Jun 16. Microbiol Spectr. 2025. PMID: 40522089 Free PMC article.
-
Cardinium disrupts Wolbachia-host dynamics in the domestic mite Tyrophagus putrescentiae: evidence from manipulative experiments.mSystems. 2025 May 20;10(5):e0176924. doi: 10.1128/msystems.01769-24. Epub 2025 Apr 18. mSystems. 2025. PMID: 40249197 Free PMC article.
-
Cardinium inhibits Wolbachia in its mite host, Tyrophagus putrescentiae, and affects host fitness.FEMS Microbiol Ecol. 2021 Sep 16;97(10):fiab123. doi: 10.1093/femsec/fiab123. FEMS Microbiol Ecol. 2021. PMID: 34448854
-
Fifty shades of bacterial endosymbionts and some of them still remain a mystery: Wolbachia and Cardinium in oribatid mites (Acari: Oribatida).J Invertebr Pathol. 2022 Mar;189:107733. doi: 10.1016/j.jip.2022.107733. Epub 2022 Feb 17. J Invertebr Pathol. 2022. PMID: 35183553 Review.
-
Horizontal Transmission of Intracellular Insect Symbionts via Plants.Front Microbiol. 2017 Nov 28;8:2237. doi: 10.3389/fmicb.2017.02237. eCollection 2017. Front Microbiol. 2017. PMID: 29234308 Free PMC article. Review.
Cited by
-
Analysis of the tripartite interactions between two bacterial symbionts, a novel Solitalea-like bacterium (Bacteroidota) and Cardinium, and the stored product mite Tyrophagus putrescentiae based on gene expression data.Microbiol Spectr. 2025 Aug 5;13(8):e0060925. doi: 10.1128/spectrum.00609-25. Epub 2025 Jun 16. Microbiol Spectr. 2025. PMID: 40522089 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
