Unique immune and other responses of human nasal epithelial cells infected with H5N1 avian influenza virus compared to seasonal human influenza A and B viruses
- PMID: 40126073
- PMCID: PMC11980200
- DOI: 10.1080/22221751.2025.2484330
Unique immune and other responses of human nasal epithelial cells infected with H5N1 avian influenza virus compared to seasonal human influenza A and B viruses
Abstract
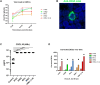
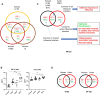
Highly pathogenic avian influenza (HPAI) virus (e.g. H5N1) infects the lower airway to cause severe infections, and constitute a prime candidate for the emergence of disease X. The nasal epithelium is the primary portal of entry for respiratory pathogens, serving as the airway's physical and immune barrier. While HPAI virus predominantly infects the lower airway, not much is known about its interactions with the nasal epithelium. Hence, we sought to elucidate and compare the differential responses of the nasal epithelium against HPAI infection that may contribute to its pathology, and to identify critical response markers. We infected human nasal epithelial cells (hNECs) cultured at the air-liquid interface from multiple healthy donors with clinical isolates of major human seasonal influenza viruses (H1N1, H3N2, influenza B) and HPAI H5N1. The infected cells were subjected to virologic, transcriptomic and secretory protein analyses. While less adapted to infecting the nasal epithelium, HPAI H5N1 elicited unique host responses unlike seasonal influenza. Interestingly, H5N1 infection of hNECs induced responses indicative of subdued antiviral activity (e.g. reduced expression of IFNβ, and inflammasome mediators, IL-1α and IL-1β); decreased wound healing; suppressed re-epithelialization; compromised epithelial barrier integrity; diminished responses to oxidative stress; and increased transmembrane solute and ion carrier gene expression. These unique molecular changes in response to H5N1 infection may represent potential targets for enhancing diagnostic and therapeutic strategies for better surveillance and management of HPAI infection in humans.
Keywords: H5N1; Influenza; highly pathogenic avian influenza; host pathogen interactions; human nasal epithelial cells; molecular responses; upper airway.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- Charostad J, Rezaei Zadeh Rukerd M, Mahmoudvand S, et al. . A comprehensive review of highly pathogenic avian influenza (HPAI) H5N1: an imminent threat at doorstep. Travel Med Infect Dis. 2023 Aug 30;55:102638. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
