Identification and Biological Evaluation of a Novel CLK4 Inhibitor Targeting Alternative Splicing in Pancreatic Cancer Using Structure-Based Virtual Screening
- PMID: 40126184
- PMCID: PMC12097107
- DOI: 10.1002/advs.202416323
Identification and Biological Evaluation of a Novel CLK4 Inhibitor Targeting Alternative Splicing in Pancreatic Cancer Using Structure-Based Virtual Screening
Abstract
Pancreatic cancer is an aggressive malignancy with a poor prognosis and limited treatment options. Cdc-like kinase 4 (CLK4), a kinase that regulates alternative splicing by phosphorylating spliceosome components, is implicated in aberrant splicing events driving pancreatic cancer progression. In this study, we established a computational model that integrates pharmacological interactions of CLK4 inhibitors with an improved hit rate. Through this model, we identified a novel CLK4 inhibitor, compound 150441, with a 50% inhibitory concentration (IC50) value of 21.4 nm. Structure-activity relationship analysis was performed to investigate key interactions and functional groups. Kinase profiling revealed that compound 150441 is selective for CLK4. Subsequent in vitro assays demonstrated that this inhibitor effectively suppressed cell growth and viability of pancreatic cancer cells. In addition, it inhibited the phosphorylation of key splicing factors, including serine- and arginine-rich splicing factor (SRSF) 4 and SRSF6. Cell cycle analysis further indicated that the compound induced G2/M arrest, leading to apoptosis. RNA-seq analysis revealed that the compound induced significant changes in alternative splicing and key biological pathways, including RNA processing, DNA replication, DNA damage, and mitosis. These findings suggest that compound 150441 has promising potential for further development as a novel pancreatic cancer treatment.
Keywords: CLK4; alternative splicing; kinase inhibitor; pancreatic cancer; structure‐based virtual screening.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
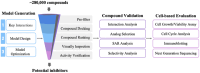
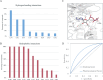
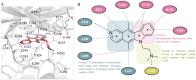
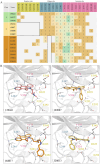



References
MeSH terms
Substances
Grants and funding
- NSTC 113-2320-B-038-009/National Science and Technology Council
- 113-2320-B-038-010/National Science and Technology Council
- 113-2320-B-400-007/National Science and Technology Council
- 113-2320-B-182A-002/National Science and Technology Council
- DP2-TMU-113-C-01/Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan
LinkOut - more resources
Full Text Sources
Medical
