The Unpaved Road of Non-Coding RNA Structure-Function Relationships: Current Knowledge, Available Methodologies, and Future Trends
- PMID: 40126344
- PMCID: PMC11932211
- DOI: 10.3390/ncrna11020020
The Unpaved Road of Non-Coding RNA Structure-Function Relationships: Current Knowledge, Available Methodologies, and Future Trends
Abstract
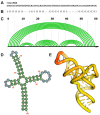
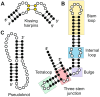
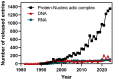
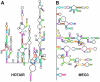
The genomes from complex eukaryotes are enriched in non-coding genes whose transcription products (non-coding RNAs) are involved in the regulation of genomic output at different levels. Non-coding RNA action is predominantly driven by sequence and structural motifs that interact with specific functional partners. Despite the exponential growth in primary RNA sequence data facilitated by next-generation sequencing studies, the availability of tridimensional RNA data is comparatively more limited. The subjacent reasons for this relative lack of information regarding RNA structure are related to the specific chemical nature of RNA molecules and the limitations of the currently available methods for structural characterization of biomolecules. In this review, we describe and analyze the different structural motifs involved in non-coding RNA function and the wet-lab and computational methods used to characterize their structure-function relationships, highlighting the current need for detailed structural studies to explore the molecular determinants of non-coding RNA function.
Keywords: X-ray crystallography; chemical probing; cryo-EM; non-coding RNA; structure–function relationships.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

