Mechanism of Action of circRNA/miRNA Network in DLBCL
- PMID: 40126346
- PMCID: PMC11932212
- DOI: 10.3390/ncrna11020022
Mechanism of Action of circRNA/miRNA Network in DLBCL
Abstract
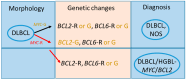
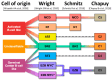
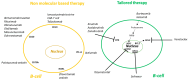
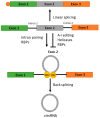
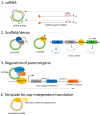
Circular RNAs (circRNAs) make up approximately 10% of the human transcriptome. CircRNAs belong to the broad group of non-coding RNAs and characteristically are formed by backsplicing into a stable circular loop. Their main role is to regulate transcription through the inhibition of miRNAs' expression, termed miRNA sponging. CircRNAs promote tumorigenesis/lymphomagenesis by competitively binding to miRNAs at miRNA binding sites. In diffuse large B-cell lymphoma (DLBCL), several circRNAs have been identified and their expression is related to both progression and response to therapy. DLBCL is the most prevalent and aggressive subtype of B-cell lymphomas and accounts for about 25% to 30% of all non-Hodgkin lymphomas. DLBCL displays great heterogeneity concerning histopathology, biology, and genetics. Patients who have relapsed or have refractory disease after first-line therapy have a very poor prognosis, demonstrating an important unmet need for new treatment options. As more circRNAs are identified in the future, we will better understand their biological roles and potential use in treating cancer, including DLBCL. For example, circAmotl1 promotes nuclear translocation of MYC and upregulation of translational targets of MYC, thus enhancing lymphomagenesis. Another example is circAPC, which is significantly downregulated in DLBCL and correlates with disease aggressiveness and poor prognosis. CircAPC increases expression of the host gene adenomatous polyposis coli (APC), and in doing so inactivates the canonical Wnt/β-catenin signaling and restrains DLBCL growth. MiRNAs belong to the non-coding regulatory molecules that significantly contribute to lymphomagenesis through their target mRNAs. In DLBCL, among the highly expressed miRNAs, are miR-155-5p and miR-21-5p, which regulate NF-ĸB and PI3K/AKT signaling pathways. The aim of this review is to describe the function and mechanism of regulation of circRNAs on miRNAs' expression in DLBCL. This will help us to better understand the regulatory network of circRNA/miRNA/mRNA, and to propose novel therapeutic targets to treat DLBCL.
Keywords: B-cells; circRNA; gene expression; lymphoma; miRNA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Crump M., Neelapu S.S., Farooq U., van den Neste E., Kuruvilla J., Westin J., Link B.K., Hay A., Cerhan J.R., Zhu L., et al. Outcomes in refractory diffuse large B-cell lymphoma: Results from the international SCHOLAR-1 study. Blood. 2017;130:1800–1808. doi: 10.1182/blood-2017-03-769620. - DOI - PMC - PubMed
-
- Chatterjee N., Hartge P., Cerhan J.R., Cozen W., Davis S., Ishibe N., Colt J., Goldin L., Severson R.K. Risk of Non-Hodgkin’s Lymphoma and Family History of Lymphatic, Hematologic, and Other Cancers. Cancer Epidemiol. Biomark. Prev. 2004;13:1415–1421. doi: 10.1158/1055-9965.1415.13.9. - DOI - PubMed
-
- Swertlow S.H., Harris L.N., Jaffe E.S. World Health Organization Classification of Tumours. WHO; Geneva, Switzerland: 2017.
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

