Genomic analysis of 11,555 probands identifies 60 dominant congenital heart disease genes
- PMID: 40127276
- PMCID: PMC12002227
- DOI: 10.1073/pnas.2420343122
Genomic analysis of 11,555 probands identifies 60 dominant congenital heart disease genes
Abstract
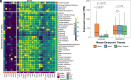
Congenital heart disease (CHD) is a leading cause of infant mortality. We analyzed de novo mutations (DNMs) and very rare transmitted/unphased damaging variants in 248 prespecified genes in 11,555 CHD probands. The results identified 60 genes with a significant burden of heterozygous damaging variants. Variants in these genes accounted for CHD in 10.1% of probands with similar contributions from de novo and transmitted variants in parent-offspring trios that showed incomplete penetrance. DNMs in these genes accounted for 58% of the signal from DNMs. Thirty-three genes were linked to a single CHD subtype while 12 genes were associated with 2 to 4 subtypes. Seven genes were only associated with isolated CHD, while 37 were associated with 1 or more extracardiac abnormalities. Genes selectively expressed in the cardiomyocyte lineage were associated with isolated CHD, while those widely expressed in the brain were also associated with neurodevelopmental delay (NDD). Missense variants introducing or removing cysteines in epidermal growth factor (EGF)-like domains of NOTCH1 were enriched in tetralogy of Fallot and conotruncal defects, unlike the broader CHD spectrum seen with loss of function variants. Transmitted damaging missense variants in MYH6 were enriched in multiple CHD phenotypes and account for ~1% of all probands. Probands with characteristic mutations causing syndromic CHD were frequently not diagnosed clinically, often due to missing cardinal phenotypes. CHD genes that were positively or negatively associated with development of NDD suggest clinical value of genetic testing. These findings expand the understanding of CHD genetics and support the use of molecular diagnostics in CHD.
Keywords: congenital heart disease; exome sequencing; genomics; human genetics; molecular inversion probes.
Conflict of interest statement
Competing interests statement:M.Y. is a co-founder and sultan to Fabric Genomics. M.Y. has stock interests exceeding $5,000 in value in Fabric Genomics.
Figures


References
-
- van der Linde D., et al. , Birth prevalence of congenital heart disease worldwide: A systematic review and meta-analysis. J. Am. Coll. Cardiol. 58, 2241–2247 (2011). - PubMed
-
- Whittemore R., Wells J. A., Castellsague X., A second-generation study of 427 probands with congenital heart defects and their 837 children. J. Am. Coll. Cardiol. 23, 1459–1467 (1994). - PubMed
MeSH terms
Substances
Grants and funding
- U01 HL128711/HL/NHLBI NIH HHS/United States
- RM1HG011014/HHS | NIH | National Human Genome Research Institute (NHGRI)
- UO1 HL128711/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- UO1 HL098147/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- U01 HL098162/HL/NHLBI NIH HHS/United States
- UO1 HL153009/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- UO1 HL098162/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- U54 HG006504/HG/NHGRI NIH HHS/United States
- UL1 TR000003/TR/NCATS NIH HHS/United States
- R00HL143036-02/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- UO1 HL131003/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- 1UG1HL135680-01/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- CDI-FR-2021-926/Children's Discovery Institute (CDI)
- NIH R03HD100883-A1/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- UG1 HL135680/HL/NHLBI NIH HHS/United States
- T32 HD007149/HD/NICHD NIH HHS/United States
- R03 HD100883/HD/NICHD NIH HHS/United States
- RM1 HG011014/HG/NHGRI NIH HHS/United States
- U01 HL098153/HL/NHLBI NIH HHS/United States
- U01 HL131003/HL/NHLBI NIH HHS/United States
- 5U54HG006504/HHS | NIH | National Human Genome Research Institute (NHGRI)
- HHMI/HHMI (HHMI)
- U01 HL153009/HL/NHLBI NIH HHS/United States
- R00 HL143036/HL/NHLBI NIH HHS/United States
- HL157653/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- UL1TR000003/HHS | NIH | National Center for Advancing Translational Sciences (NCATS)
- 19PRE3438084/American Heart Association (AHA)
- K08 HL157653/HL/NHLBI NIH HHS/United States
- U01 HL098147/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

