The FBXW7-KMT2 axis in cancer-associated fibroblasts controls tumor growth via an epigenetic-paracrine mechanism
- PMID: 40127278
- PMCID: PMC12002300
- DOI: 10.1073/pnas.2423130122
The FBXW7-KMT2 axis in cancer-associated fibroblasts controls tumor growth via an epigenetic-paracrine mechanism
Abstract
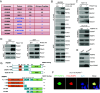
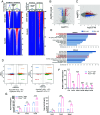
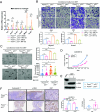
F-box and WD repeat domain-containing 7 (FBXW7) is a tumor suppressor that targets various oncoproteins for degradation, but its role in modulating cancer-associated fibroblasts (CAFs) in the tumor microenvironment remains elusive. Here, we report that FBXW7 expression is gradually downregulated in CAFs during the progression of human pancreatic and lung cancers. Mechanically, FBXW7 inhibits histone lysine methyltransferase 2 (KMT2) methyltransferase activity via retinoblastoma binding protein 5 (RbBP5) binding, whereas FBXW7 depletion abrogates the binding to activate KMT2, leading to increased H3K4 methylations and global upregulation of gene expression. Activation of the interleukin-17 (IL-17) signaling pathway triggers the secretion of cytokines and chemokines to promote migration, invasion, and sphere formation of lung cancer cells. Coinjection of Fbxw7-depleted mouse embryonic fibroblasts with cancer cells enhances in vivo tumor growth, demonstrating a paracrine effect. Hypoxia downregulates CAF FBXW7 via ETS proto-oncogene 1 (ETS1) to increase H3K4 methylation, whereas conditioned media from hypoxia-exposed CAFs promotes migration and invasion of pancreatic cancer cells, highlighting FBXW7's tumor-suppressing role through KMT2 inactivation.
Keywords: FBXW7 E3 ligase; IL-17 signaling pathway; epigenetic regulation; methyltransferase; paracrine effect.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- de Visser K. E., Joyce J. A., The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 41, 374–403 (2023). - PubMed
-
- Erez N., Truitt M., Olson P., Arron S. T., Hanahan D., Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 17, 135–147 (2010). - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFC3401500/MOST | National Key Research and Development Program of China (NKPs)
- 2021YFA1101000/MOST | National Key Research and Development Program of China (NKPs)
- LD22H300003/MOST | NSFC | NSFC-Zhejiang Joint Fund | | Natural Science Foundation of Zhejiang Province (ZJNSF)
- 82188102/MOST | National Natural Science Foundation of China (NSFC)
- 92253203/MOST | National Natural Science Foundation of China (NSFC)
- U22A20317/MOST | National Natural Science Foundation of China (NSFC)
- 81974429/MOST | National Natural Science Foundation of China (NSFC)
- 82172898/MOST | National Natural Science Foundation of China (NSFC)
- 32170656/MOST | National Natural Science Foundation of China (NSFC)
- Z211100002121039/åŒ-京å¸'ç§'å¦æŠ€æœ¯å§"å'˜ä¼š | Beijing Nova Program
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

