Spt6-Spn1 interaction is required for RNA polymerase II association and precise nucleosome positioning along transcribed genes
- PMID: 40127868
- PMCID: PMC12053661
- DOI: 10.1016/j.jbc.2025.108436
Spt6-Spn1 interaction is required for RNA polymerase II association and precise nucleosome positioning along transcribed genes
Abstract
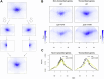
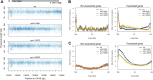
Spt6-Spn1 is an essential histone chaperone complex that associates with RNA Polymerase II (RNAPII) and reassembles nucleosomes during gene transcription. While the interaction between Spt6 and Spn1 is important for its histone deposition and transcription functions, a precise mechanistic understanding is still limited. Here, using temperature-sensitive alleles of spt6 and spn1 that disrupt their interaction in yeast, we show that the Spt6-Spn1 association is important for its stable interaction with the elongating RNAPII complex and nucleosomes. Using micrococcal nuclease (MNase)-based chromatin occupancy profiling, we further find that Spt6-Spn1 interaction is required to maintain a preferred nucleosome positioning at actively transcribed genes; in the absence of Spt6-Spn1 interaction, we observe a return to replication-dependent phasing. In addition to positioning defects, Spt6-Spn1 disrupting mutants also resulted in an overall shift of nucleosomes toward the 5' end of genes that were correlated with decreased RNAPII levels. As loss of Spt6-Spn1 association results in cryptic transcription at a subset of genes, we examined these genes for their nucleosome profiles. These findings revealed that the chromatin organization at these loci is similar to other active genes, thus underscoring the critical role of DNA sequence in mediating cryptic transcription when nucleosome positioning is altered. Taken together, these findings reveal that Spt6-Spn1 interaction is key to its association with elongating RNAPII and to its ability to precisely organize nucleosomes across transcription units.
Keywords: chromatin; histone chaperone; histones; nucleosomes; spn1; spt6; transcription; yeast.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests. BDS is a co-founder and board member of EpiCypher, Inc. BDS is an Editorial Associate Editor for Journal of Biological Chemistry and was not involved in the editorial review or the decision to publish this article.
Figures


References
-
- Gurard-Levin Z.A., Quivy J.P., Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014;83:487–517. - PubMed
-
- Ray-Gallet D., Almouzni G. H3-H4 histone chaperones and cancer. Curr. Opin. Genet. Dev. 2022;73 - PubMed
-
- Aoi Y., Shilatifard A. Transcriptional elongation control in developmental gene expression, aging, and disease. Mol. Cell. 2023;83:3972–3999. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

