MRN-CtIP, EXO1, and DNA2-WRN/BLM act bidirectionally to process DNA gaps in PARPi-treated cells without strand cleavage
- PMID: 40127955
- PMCID: PMC12047661
- DOI: 10.1101/gad.352421.124
MRN-CtIP, EXO1, and DNA2-WRN/BLM act bidirectionally to process DNA gaps in PARPi-treated cells without strand cleavage
Abstract
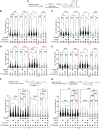
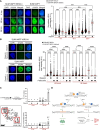
Single-stranded DNA (ssDNA) gaps impact genome stability and PARP inhibitor (PARPi) sensitivity, especially in BRCA1/2-deficient tumors. Using single-molecule DNA fiber analysis, electron microscopy, and biochemical methods, we found that MRN, CtIP, EXO1, and DNA2-WRN/BLM resect ssDNA gaps through a mechanism different from their actions at DNA ends. MRN resects ssDNA gaps in the 3'-to-5' direction using its pCtIP-stimulated exonuclease activity. Unlike at DNA ends, MRN does not use its endonucleolytic activity to cleave the 5'-terminated strand flanking the gap or the ssDNA. EXO1 and DNA2-WRN/BLM specifically resect the 5' end of the gap independent of MRN-CtIP. This resection process alters ssDNA gap repair kinetics in BRCA1-proficient and -deficient cells. In BRCA1-deficient cells treated with PARPis, excessive resection results in larger ssDNA gaps, hindering their repair and leading to DNA breaks in subsequent cell cycle stages due to ssDNA gaps colliding with DNA replication forks. These findings broaden our understanding of the role of human nucleases in DNA metabolism and have significant implications for defining the mechanisms driving PARPi sensitivity in BRCA-deficient tumors.
Keywords: BRCA; DNA replication; DNA replication stress; MRE11; PARP inhibitor; genome stability; nucleases; single-stranded DNA gaps.
© 2025 Seppa et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
