Targeting the disrupted Hippo signaling to prevent neoplastic renal epithelial cell immune evasion
- PMID: 40128178
- PMCID: PMC11933345
- DOI: 10.1038/s41467-025-57697-7
Targeting the disrupted Hippo signaling to prevent neoplastic renal epithelial cell immune evasion
Erratum in
-
Author Correction: Targeting the disrupted Hippo signaling to prevent neoplastic renal epithelial cell immune evasion.Nat Commun. 2025 Jun 30;16(1):5415. doi: 10.1038/s41467-025-61430-9. Nat Commun. 2025. PMID: 40588475 Free PMC article. No abstract available.
Abstract
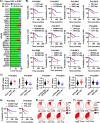
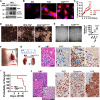
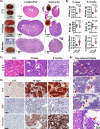
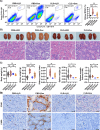
Large-scale cancer genetic/genomic studies demonstrated that papillary renal cell carcinoma (pRCC) is featured with a frequent shallow deletion of the upstream tumor suppressors of the Hippo/YAP signaling pathway, suggesting that this signaling pathway may play a role in pRCC development. Here we develop a transgenic mouse model with a renal epithelial cell-specific hyperactivation of YAP1 and find that hyperactivation of YAP1 can induce dedifferentiation and transformation of renal tubular epithelial cells leading to the development of pRCC. We analyze at the single-cell resolution the cellular landscape alterations during cancer initiation and progression. Our data indicate that the hyperactivated YAP1, via manipulating multiple signaling pathways, induces epithelial cell transformation, MDSC (Myeloid-derived suppressor cells) accumulation, and pRCC development. Interestingly, we find that depletion of MDSC blocks YAP1-induced kidney overgrowth and tumorigenesis. Inhibiting YAP1 activity with MGH-CP1, a recently developed TEAD inhibitor, impedes MDSC accumulation and suppresses tumor development. Our results identify the disrupted Hippo/YAP signaling as a major contributor to pRCC and suggest that targeting the disrupted Hippo pathway represents a plausible strategy to prevent and treat pRCC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Dr. Xu Wu has a financial interest in Tasca Therapeutics, which is developing small molecule modulators of TEAD palmitoylation and transcription factors. Dr. Wu’s interests were reviewed and are managed by Mass General Hospital, and Mass General Brigham in accordance with their conflict-of-interest policies. The rest authors have no competing interests to declare.
Figures




References
-
- Harvey, K. & Tapon, N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat. Rev. Cancer7, 182–191 (2007). - PubMed
MeSH terms
Substances
Grants and funding
- 1R01CA201500/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA197976/CA/NCI NIH HHS/United States
- 1R0CA1238270/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA201500/CA/NCI NIH HHS/United States
- R01 CA279385/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

