S-palmitoylation modulates ATG2-dependent non-vesicular lipid transport during starvation-induced autophagy
- PMID: 40128367
- PMCID: PMC12048663
- DOI: 10.1038/s44318-025-00410-7
S-palmitoylation modulates ATG2-dependent non-vesicular lipid transport during starvation-induced autophagy
Abstract
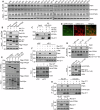
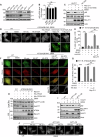
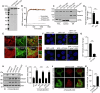
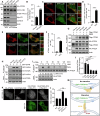
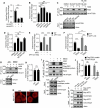
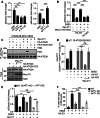
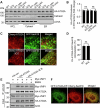
Lipid transfer proteins mediate the non-vesicular transport of lipids at membrane contact sites to regulate the lipid composition of organelle membranes. Despite significant recent advances in our understanding of the structural basis for lipid transfer, its functional regulation remains unclear. In this study, we report that S-palmitoylation modulates the cellular function of ATG2, a rod-like lipid transfer protein responsible for transporting phospholipids from the endoplasmic reticulum (ER) to phagophores during autophagosome formation. During starvation-induced autophagy, ATG2A undergoes depalmitoylation as the balance between ZDHHC11-mediated palmitoylation and APT1-mediated depalmitoylation. Inhibition of ATG2A depalmitoylation leads to impaired autophagosome formation and disrupted autophagic flux. Further, in cell and in vitro analyses demonstrate that S-palmitoylation at the C-terminus of ATG2A anchors the C-terminus to the ER. Depalmitoylation detaches the C-terminus from the ER membrane, enabling it to interact with phagophores and promoting their growth. These findings elucidate a S-palmitoylation-dependent regulatory mechanism of cellular ATG2, which may represent a broad regulatory strategy for lipid transport mediated by bridge-like transporters within cells.
Keywords: ATG2; Autophagy; Lipid Transfer Protein; S-palmitoylation.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

