Epigenetic editing at individual age-associated CpGs affects the genome-wide epigenetic aging landscape
- PMID: 40128456
- PMCID: PMC12176646
- DOI: 10.1038/s43587-025-00841-1
Epigenetic editing at individual age-associated CpGs affects the genome-wide epigenetic aging landscape
Abstract
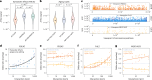
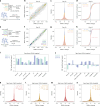
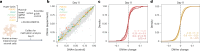
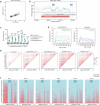
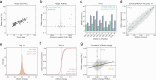
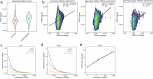
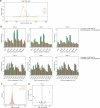
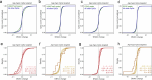
Aging is reflected by genome-wide DNA methylation changes, which form the basis of epigenetic clocks, but it is largely unclear how these epigenetic modifications are regulated and whether they directly affect the aging process. In this study, we performed epigenetic editing at age-associated CpG sites to explore the consequences of interfering with epigenetic clocks. CRISPR-guided editing targeted at individual age-related CpGs evoked genome-wide bystander effects, which were highly reproducible and enriched at other age-associated regions. 4C-sequencing at age-associated sites revealed increased interactions with bystander modifications and other age-related CpGs. Subsequently, we multiplexed epigenetic editing in human T cells and mesenchymal stromal cells at five genomic regions that become either hypermethylated or hypomethylated upon aging. While targeted methylation seemed more stable at age-hypermethylated sites, both approaches induced bystander modifications at CpGs with the highest correlations with chronological age. Notably, these effects were simultaneously observed at CpGs that gain and lose methylation with age. Our results demonstrate that epigenetic editing can extensively modulate the epigenetic aging network and interfere with epigenetic clocks.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: W.W. is cofounder of the company Cygenia ( www.cygenia.com ), which provides services for epigenetic analyses to other scientists. The other authors declare no competing interests.
Figures






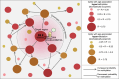
References
-
- Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet.19, 371–384 (2018). - PubMed
MeSH terms
Grants and funding
- WA 1706/12-2 within CRU344/417911533; WA1706/14-1; SFB 1506/1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- ForTra (PrickMe)/Else Kröner-Fresenius-Stiftung (Else Kroner-Fresenius Foundation)
- VIP+: 03VP11580/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
- thesis stipend/José Carreras Leukämie-Stiftung (Deutsche José Carreras Leukämie-Stiftung)
LinkOut - more resources
Full Text Sources
Medical

