Benchmarking metagenomic binning tools on real datasets across sequencing platforms and binning modes
- PMID: 40128535
- PMCID: PMC11933696
- DOI: 10.1038/s41467-025-57957-6
Benchmarking metagenomic binning tools on real datasets across sequencing platforms and binning modes
Abstract
Metagenomic binning is a culture-free approach that facilitates the recovery of metagenome-assembled genomes by grouping genomic fragments. However, there remains a lack of a comprehensive benchmark to evaluate the performance of metagenomic binning tools across various combinations of data types and binning modes. In this study, we benchmark 13 metagenomic binning tools using short-read, long-read, and hybrid data under co-assembly, single-sample, and multi-sample binning, respectively. The benchmark results demonstrate that multi-sample binning exhibits optimal performance across short-read, long-read, and hybrid data. Moreover, multi-sample binning outperforms other binning modes in identifying potential antibiotic resistance gene hosts and near-complete strains containing potential biosynthetic gene clusters across diverse data types. This study also recommends three efficient binners across all data-binning combinations, as well as high-performance binners for each combination.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors declare no competing interests.
Figures

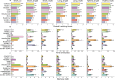

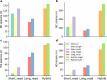
References
-
- Jansson, J. K. Microorganisms, climate change, and the sustainable development goals: progress and challenges. Nat. Rev. Microbiol.21, 622–623 (2023). - PubMed
-
- Prosser, J. I. et al. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol.5, 384–392 (2007). - PubMed
-
- Tringe, S. G. & Rubin, E. M. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet.6, 805–814 (2005). - PubMed
-
- Albertsen, M. Long-read metagenomics paves the way toward a complete microbial tree of life. Nat. Methods20, 30–31 (2023). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

