Metagenomic signatures of extraintestinal bacterial infection in the febrile term infant gut microbiome
- PMID: 40128855
- PMCID: PMC11931804
- DOI: 10.1186/s40168-025-02079-w
Metagenomic signatures of extraintestinal bacterial infection in the febrile term infant gut microbiome
Abstract
Background: Extraintestinal bacterial infections (EBIs), e.g., urinary tract infection, bacteremia, and meningitis, occur in approximately 10% of febrile infants younger than 60 days. Although many EBI-causing species commonly reside in the infant gut, proof that the digestive system is a pre-infection habitat remains unestablished.
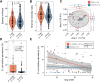
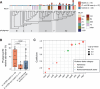
Results: We studied a cohort of febrile term infants < 60 days old who presented to one of thirteen US emergency departments in the Pediatric Emergency Care Applied Research Network from 2016 to 2019. Forty EBI cases and 74 febrile controls matched for age, sex, and race without documented EBIs were selected for analysis. Shotgun sequencing was performed of the gut microbiome and of strains cultured from the gut and extraintestinal site(s) of EBI cases, including blood, urine, and/or cerebrospinal fluid. Using a combination of EBI isolate genomics and fecal metagenomics, we detected an intestinal strain presumptively isogenic to the EBI pathogen (> 99.999% average nucleotide identity) in 63% of infants with EBIs. Although there was no difference in gut microbiome diversity between cases and controls, we observed significantly increased Escherichia coli relative abundance in the gut microbiome of infants with EBIs caused by E. coli. Infants with E. coli infections who were colonized by the putatively isogenic pathogen strain had significantly higher E. coli phylogroup B2 abundance in their gut, and their microbiome was more likely to contain virulence factor loci associated with adherence, exotoxin production, and nutritional/metabolic function.
Conclusions: The intestine plausibly serves as a reservoir for EBI pathogens in a subset of febrile term infants, prompting consideration of new opportunities for surveillance and EBI prevention among colonized, pre-symptomatic infants. Video Abstract.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the institutional review board at all sites. Secondary analyses conducted here were approved under Washington University HRPO IRB#202106080. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Greenhow TL, Hung Y-Y, Herz AM, Losada E, Pantell RH. The changing epidemiology of serious bacterial infections in young infants. Pediatr Infect Dis J. 2014;33:595–9. - PubMed

