Rapid serotype-independent differential detection of biofilm-positive and biofilm-negative Salmonella using Fourier transform infrared biotyping
- PMID: 40129480
- PMCID: PMC11931386
- DOI: 10.1016/j.onehlt.2025.101004
Rapid serotype-independent differential detection of biofilm-positive and biofilm-negative Salmonella using Fourier transform infrared biotyping
Abstract
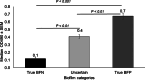
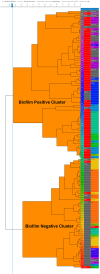
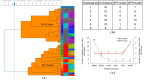
Foodborne illnesses caused by Salmonella represent a global one health challenge, with biofilm-forming strains exhibiting enhanced public health risks due to their ability to persist due to resistance to antimicrobial agents, disinfectants, and environmental stresses. While food-safety and public health investigation primarily focus on Salmonella identification and source tracing, they often overlook the biofilm-forming capacity of isolates, limiting their predictive value for risks posed by biofilm producing Salmonella. This study assessed fourier transform infrared (FTIR) biotyping for rapid serotype-independent differentiatial detection of biofilm-positive (BFP) from biofilm-negative (BFN) Salmonella. A total of 270 Salmonella strains representing 12 common serotypes were classified using three conventional biofilm assays (congo red and coomassie brilliant blue agar test, calcofluor test, and tube test) into true BFP (n = 80), true BFN (n = 64), and uncertain (n = 59) biofilm producers. Biofilm production for each group was also assessed with a microtiter plate assay. FTIR biotyping was applied to a subset of 115 strains (61 BFP, 54 BFN). Using spectral windows of 1180-1050 cm-1 and 1400-1200 cm-1, FTIR biotyping accurately differentiated BFP from BFN strains with 93.4 % sensitivity, 83.3 % specificity, and 88.6 % overall accuracy. FTIR biotyping differentiated 59 strains with uncertain biofilm status into BFN (n = 45) and BFP (n = 14). FTIR biotyping provides a rapid, sensitive and specific method for detection of biofilm-forming Salmonella strains. Incorporating FTIR biotyping for biofilm detection in current Salmonella surveillance and source-tracing protocols can enhance food safety risk assessments and improve Salmonella outbreak prevention.
Keywords: Biofilm; FTIR spectroscopy; IR Biotyper; Salmonella.
© 2025 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Classification of Salmonella enterica of the (Para-)Typhoid Fever Group by Fourier-Transform Infrared (FTIR) Spectroscopy.Microorganisms. 2021 Apr 15;9(4):853. doi: 10.3390/microorganisms9040853. Microorganisms. 2021. PMID: 33921159 Free PMC article.
-
Phenotypic and genotypic characterization of antimicrobial resistance and virulence profiles of Salmonella enterica serotypes isolated from necropsied horses in Kentucky.Microbiol Spectr. 2025 Mar 4;13(3):e0250124. doi: 10.1128/spectrum.02501-24. Epub 2025 Jan 23. Microbiol Spectr. 2025. PMID: 39846771 Free PMC article.
-
Machine learning-based typing of Salmonella enterica O-serogroups by the Fourier-Transform Infrared (FTIR) Spectroscopy-based IR Biotyper system.J Microbiol Methods. 2022 Oct;201:106564. doi: 10.1016/j.mimet.2022.106564. Epub 2022 Sep 6. J Microbiol Methods. 2022. PMID: 36084763
-
A novel MIR imaging approach for precise detection of S. epidermidis biofilms in seconds.Biofilm. 2025 Mar 6;9:100270. doi: 10.1016/j.bioflm.2025.100270. eCollection 2025 Jun. Biofilm. 2025. PMID: 40130066 Free PMC article. Review.
-
Biofilm formation in food processing plants and novel control strategies to combat resistant biofilms: the case of Salmonella spp.Food Sci Biotechnol. 2023 May 30;32(12):1703-1718. doi: 10.1007/s10068-023-01349-3. eCollection 2023 Oct. Food Sci Biotechnol. 2023. PMID: 37780596 Free PMC article. Review.
References
-
- Burden of Foodborne Illness: Findings | Estimates of Foodborne Illness | CDC 2023. https://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html (accessed April 12, 2024)
-
- Painter J.A., Hoekstra R.M., Ayers T., Tauxe R.V., Braden C.R., Angulo F.J., et al. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 2013;19:407–415. doi: 10.3201/eid1903.111866. - DOI - PMC - PubMed
-
- Steenackers H., Hermans K., Vanderleyden J., De Keersmaecker S.C.J. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012;45:502–531. doi: 10.1016/j.foodres.2011.01.038. - DOI
LinkOut - more resources
Full Text Sources

