Targeting Piezo1 channel to alleviate intervertebral disc degeneration
- PMID: 40129609
- PMCID: PMC11930658
- DOI: 10.1016/j.jot.2025.01.006
Targeting Piezo1 channel to alleviate intervertebral disc degeneration
Abstract
Background: Low back pain impacts over 600 million people worldwide, predominantly due to intervertebral disc degeneration. This study focuses on the role of Piezo1, a crucial mechanosensitive ion channel protein, in the pathology and potential treatment of disc degeneration.
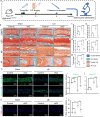
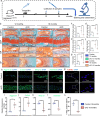
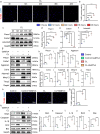
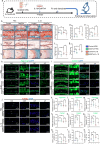
Materials and methods: To investigate the effects of disc-specific Piezo1 deletion, we generated Aggrecan CreERT2 ; Piezo1 fl/fl mice and examined both lumbar spine instability (LSI)- and aging-induced disc degeneration. Additionally, the effect of pharmacological inhibition of Piezo1 was evaluated using GsMTx4, a potent Piezo1 antagonist, in an ex vivo model stimulated with IL-1β to induce disc degeneration. Assessments included histological examinations, immunofluorescence, and western blot analyses to thoroughly characterize the alterations in the intervertebral discs.
Results: Elevated expression of Piezo1 was detected in the nucleus pulposus (NP) of intervertebral discs with advanced disc degeneration in both aged mice and human patients. Inducible deletion of Piezo1 expression in aggrecan-expressing disc cells significantly reduced lumbar disc degeneration, decreased extracellular matrix (ECM) degradation, and lowered apoptosis in NP cells, observed in both aged mice and those undergoing LSI surgery. Excessive compression loading (CL) upregulated Piezo1 expression, induced ECM disruption, and increased apoptosis in NP cells, whereas inhibition of Piezo1 with GsMTx4 effectively mitigated these pathological changes. Furthermore, in ex vivo cultured mouse discs, GsMTx4 treatment significantly alleviated IL-1β-induced degenerative damages, restored ECM anabolism, and reduced apoptosis.
Conclusions: The findings suggest that Piezo1 plays a critical role in the development of disc degeneration and highlight its potential as a therapeutic target. Inhibiting Piezo1 could offer a novel strategy for treating or preventing this critical disease.
Translational potential of this article: This research highlights the involvement of Piezo1 in the development of intervertebral disc degeneration and emphasizes the potential for targeting Piezo1 as a therapeutic strategy to delay or reverse this condition.
Keywords: Aging; GsMTx4; Intervertebral disc degeneration; Lumbar spine instability; Piezo1.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures



Similar articles
-
Excessive mechanical stress-induced intervertebral disc degeneration is related to Piezo1 overexpression triggering the imbalance of autophagy/apoptosis in human nucleus pulpous.Arthritis Res Ther. 2022 May 23;24(1):119. doi: 10.1186/s13075-022-02804-y. Arthritis Res Ther. 2022. PMID: 35606793 Free PMC article.
-
Aberrant mechanical loading induces annulus fibrosus cells apoptosis in intervertebral disc degeneration via mechanosensitive ion channel Piezo1.Arthritis Res Ther. 2023 Jul 7;25(1):117. doi: 10.1186/s13075-023-03093-9. Arthritis Res Ther. 2023. PMID: 37420255 Free PMC article.
-
Mechanosensitive Ion Channel Piezo1 Activated by Matrix Stiffness Regulates Oxidative Stress-Induced Senescence and Apoptosis in Human Intervertebral Disc Degeneration.Oxid Med Cell Longev. 2021 Feb 10;2021:8884922. doi: 10.1155/2021/8884922. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 33628392 Free PMC article.
-
Role of the mechanosensitive piezo1 channel in intervertebral disc degeneration.Clin Physiol Funct Imaging. 2023 Mar;43(2):59-70. doi: 10.1111/cpf.12798. Epub 2022 Dec 8. Clin Physiol Funct Imaging. 2023. PMID: 36400723 Review.
-
Roles of large aggregating proteoglycans in human intervertebral disc degeneration.Connect Tissue Res. 2019 May;60(3):209-218. doi: 10.1080/03008207.2018.1499731. Epub 2018 Aug 9. Connect Tissue Res. 2019. PMID: 29992840 Review.
Cited by
-
Tackling musculoskeletal disorders in an aging world: Advances in orthopaedic translational research.J Orthop Translat. 2025 Apr 18;51:A1-A3. doi: 10.1016/j.jot.2025.04.003. eCollection 2025 Mar. J Orthop Translat. 2025. PMID: 40584014 Free PMC article. No abstract available.
References
-
- Roberts S., Evans H., Trivedi J., Menage J. Histology and pathology of the human intervertebral disc. JBJS. 2006;88a(suppl_2):10–14. - PubMed
-
- Kepler C.K., Ponnappan R.K., Tannoury C.A., Risbud M.V., Anderson D.G. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13(3):318–330. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

