Variants in WASHC3, a component of the WASH complex, cause short stature, variable neurodevelopmental abnormalities, and distinctive facial dysmorphism
- PMID: 40129681
- PMCID: PMC11932664
- DOI: 10.1016/j.gimo.2024.101915
Variants in WASHC3, a component of the WASH complex, cause short stature, variable neurodevelopmental abnormalities, and distinctive facial dysmorphism
Abstract
Purpose: Genetic defects that impair growth plate chondrogenesis cause a phenotype that varies from skeletal dysplasia to mild short stature with or without other syndromic features. In many individuals with impaired skeletal growth, the genetic causes remain unknown.
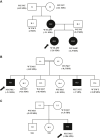
Method: Exome sequence was performed in 3 unrelated families with short stature, distinctive facies, and neurodevelopmental abnormalities. The impact of identified variants was studied in vitro.
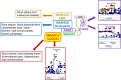
Results: Exome sequencing identified variants in WASHC3, a component of the WASH complex. In the first family, a de-novo-dominant missense variant (p.L69F) impaired WASHC3 participation in the WASH complex, altered PTH1R endosomal trafficking, diminished PTH1R signaling, and affected growth plate chondrocyte hypertrophic differentiation, providing a likely explanation for the short stature. Knockdown of other WASH complex components also diminished PTH1R signaling. In the second and third families, a homozygous variant in the start codon (p.M1?) markedly reduced WASHC3 protein expression.
Conclusion: In combination with prior studies of WASH complex proteins, our findings provide evidence that the WASH complex is required for normal skeletal growth and that, consequently, genetic abnormalities impairing the function of the WASH complex (WASHopathy) cause short stature, as well as distinctive facies and variable neurodevelopmental abnormalities.
Keywords: Growth; Neurodevelopmental abnormalities; Receptor trafficking; WASH complex; WASHC3.
© 2025 Published by Elsevier Inc. on behalf of American College of Medical Genetics and Genomics.
Conflict of interest statement
James R. Lupski has stock ownership in 23andMe, is a paid consultant for Regeneron Genetics Center, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, genomic disorders, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics (BG); James R. Lupski serves on the Scientific Advisory Board (SAB) of BG.
Figures





References
LinkOut - more resources
Full Text Sources

