Genomic islands and molecular mechanisms relating to drug-resistance in Clostridioides (Clostridium) difficile PCR ribotype 176
- PMID: 40130321
- PMCID: PMC11983580
- DOI: 10.1080/22221751.2025.2482698
Genomic islands and molecular mechanisms relating to drug-resistance in Clostridioides (Clostridium) difficile PCR ribotype 176
Abstract
Objectives: To analyse characteristics of Clostridioides difficile PCR ribotype 176 clinical isolates from Poland, the Czech Republic and Slovakia with regard to the differences in its epidemiology.
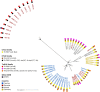
Methods: Antimicrobial susceptibility testing and whole genome sequencing were performed on a selected group of 22 clonally related isolates as determined by multilocus variable-number tandem repeat analysis (n = 509). Heterologous expression and functional analysis of the newly identified methyltransferase were performed.
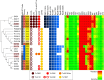
Results: Core genome multilocus sequence typing found 10-37 allele differences. All isolates were resistant to fluoroquinolones (gyrA_p. T82I), aminoglycosides with aac(6')-Ie-aph(2'')-Ia in six isolates. Erythromycin resistance was detected in 21/22 isolates and 15 were also resistant to clindamycin with ermB gene. Fourteen isolates were resistant to rifampicin with rpoB_p. R505K or p. R505K/H502N, and five to imipenem with pbp1_p. P491L and pbp3_p. N537K. PnimBG together with nimB_p. L155I were detected in all isolates but only five were resistant to metronidazole on chocolate agar. The cfrE, vanZ1 and cat-like genes were not associated with linezolid, teicoplanin and chloramphenicol resistance, respectively. The genome comparison identified six transposons carrying antimicrobial resistance genes. The ermB gene was carried by new Tn7808, Tn6189 and Tn6218-like. The aac(6')-Ie-aph(2'')-Ia were carried by Tn6218-like and new Tn7806 together with cfrE gene. New Tn7807 carried a cat-like gene. Tn6110 and new Tn7806 contained an RlmN-type 23S rRNA methyltransferase, designated MrmA, associated with high-level macrolide resistance in isolates without ermB gene.
Conclusions: Multidrug-resistant C. difficile PCR ribotype 176 isolates carry already described and unique transposons. A novel mechanism for erythromycin resistance in C. difficile was identified.
Keywords: Clostridioides difficile infection; epidemiology; macrolide resistance methyltransferase; whole genome sequencing.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Viprey VF, Davis GL, Benson AD, et al. . A point-prevalence study on community and inpatient Clostridioides difficile infections (CDI): results from combatting bacterial resistance in Europe CDI (COMBACTE-CDI), July to November 2018. Euro Surveill. 2022;27(26):2100704, doi:10.2807/1560-7917.ES.2022.27.26.2100704 - DOI - PMC - PubMed
-
- European Centre for Disease Prevention and Control . Study protocol for a survey of whole genome sequencing of Clostridioides difficile isolates from tertiary acute care hospitals, EU/EEA, 2022–2023. Stockholm: ECDC; 2024.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
