Molecular Stiffening by Macrocycle Clustering
- PMID: 40130680
- PMCID: PMC12105694
- DOI: 10.1002/anie.202420880
Molecular Stiffening by Macrocycle Clustering
Abstract
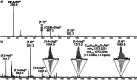
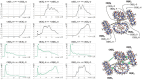
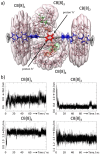
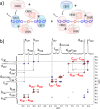
Allosteric stiffening of a portion of a protein surface is a strategy used in nature to regulate protein oligomerization and provide crucial functions for cells. However, a similar strategy to selectively control part of a compound dynamics remains elusive. Here we show that cucurbit[n]uril (CB[n]) macrocycles can bind almost all portions of a tetratopic guest molecule, stiffening the different parts of the guest to different extents. "Host-guest" interactions were found to be instrumental in selectively "freezing" guest molecular motions. The combination of 1H-NMR (1D, 2D), DOSY, VT-NMR, isothermal titration calorimetry (ITC), mass spectrometry and molecular modelling enabled to highlight the crucial role of cucurbit[8]uril (CB[8]) binding in the selective hardening of relevant portions of the guest molecule. Beyond implications for bioinspired systems mimicking control of a system dynamic to create a new function, this approach has relevance for improving room temperature phosphorescence, and could also be used to allosterically control organocatalysis in water.
Keywords: Cucurbituril; Macrocycles; Rigidification; Supramolecular; Tetratopic.
© 2025 The Author(s). Angewandte Chemie International Edition published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

